- Title
-
Dnmt1 is required for the development of auditory organs via cell cycle arrest and Fgf signalling
- Authors
- Tang, D., Zheng, S., Zheng, Z., Liu, C., Zhang, J., Yan, R., Wu, C., Zuo, N., Wu, L., Xu, H., Liu, S., He, Y.
- Source
- Full text @ Cell Prolif.
|
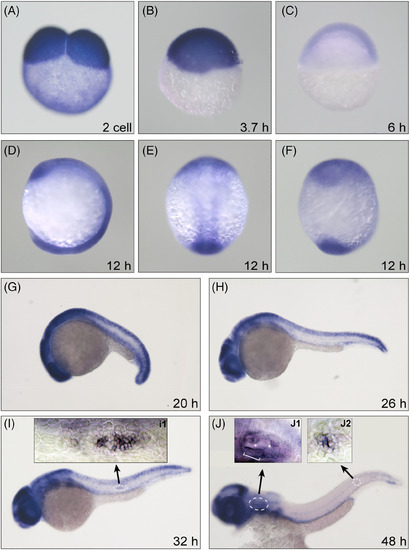
Dnmt1 is required for the development of zebrafish inner ear and pLL. The WISH staining delineates a persistent expression of Dnmt1 at two-cell stage (A), oblong stage (B), shield stage (C), segmentation period (D: lateral view, E: dorsal view, F: ventral view at 12 hpf and G: lateral view at 20 hpf). The high expression levels of Dnmt1 are concentrated on the migrating primordium (H and I), inner ear in the head region and deposited neuromasts in the posterior lateral line (J) from 26 to 48 hpf. The magnification-times images of primordium (I1), inner ear (J1) and neuromast (J2) structure indicate a strong expression of Dnmt1. The white bracket outlines sensory maculae HCs and white asterisks label semicircular canals (J1)
|
|
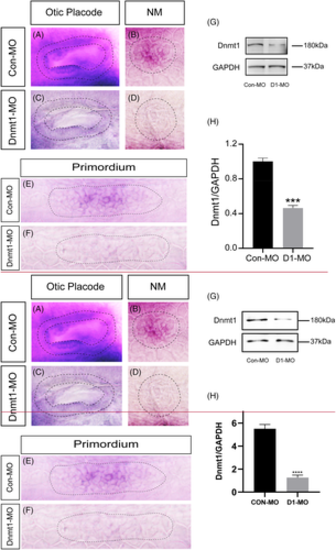
The expression of dnmt1 is markedly downregulated after knocking down of Dnmt1. A-F, In situ staining of dnmt1 is uniformly downregulated in Dnmt1-MO embryos compared to the controls by WISH at different stages. The black dotted lines outline the otic vesicle (A, C) and neuromast (B, D) at 48 hpf respectively. The black dotted lines in E-F outline the primordium at 32 hpf. G-H, The protein blotting of Dnmt1 is significantly decreased by Dnmt1 morpholino injection both in the band intensity (G) and the quantification analysis (H)
|
|
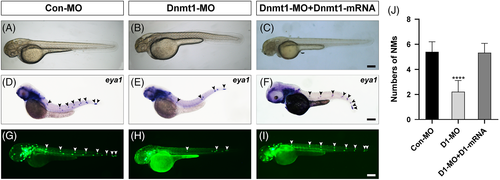
Dnmt1 is required for neuromast deposition in zebrafish posterior lateral line. A-B, At 48 hpf, the gross morphology of wild-type embryos and Dnmt1-deficient embryos. D-E, Reduced number of neuromasts and extended distance between neuromasts are found in the staining of eya1 in lateral line neuromasts when compared the controls (D) with the Dnmt1 morphants (E). G-H, The transgenic zebrafish line cldnb:lynGFP confirms the results observed in WISH. (C, F, I) Successful rescue of morphology, eya1 staining and green fluorescence labelled neuromasts are achieved in mRNA and MO co-injection groups. The black arrowheads in D-F, and the white arrowheads in G-I both mark the PLL neuromasts. Scale bars mark the 200 μm scale. (J) Statistic analysis of the number of posterior LL neuromasts at 48 hpf in controls (n = 117), Dnmt1-deficient embryos (n = 159) and Dnmt1-MO + mRNA members (n = 159). ****p < 0.0001
|
|
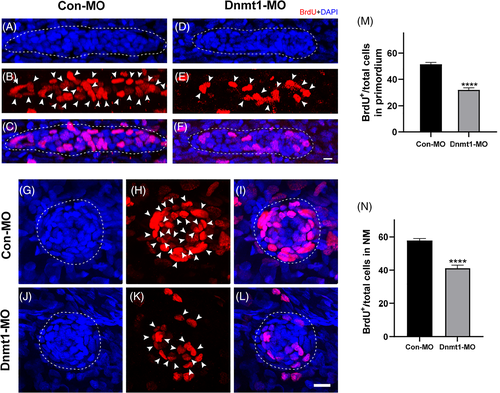
Downregulated Dnmt1 represses cell proliferation during primordium migration and neuromast formation. A-L, Representative images show the comparison in number of BrdU-labelled proliferating cells between controls embryos and Dnmt1-deficient mutants at 32 hpf in primordium (A-F) and at 48 hpf (G-L) in neuromasts. Scale bars mark the 10 μm scale. M-N, Significant differences in quantification of BrdU index in control embryos (n = 32) and Dnmt1-MO embryos (n = 31) were labelled. The arrowheads in B, E, H, K mark the proliferative cells. Dotted lines in A, C, D, F outline the primordium, and dotted lines in G, J, I, L outline the neuromast. Data are recorded as mean ± SEM. ****p < 0.0001
|
|
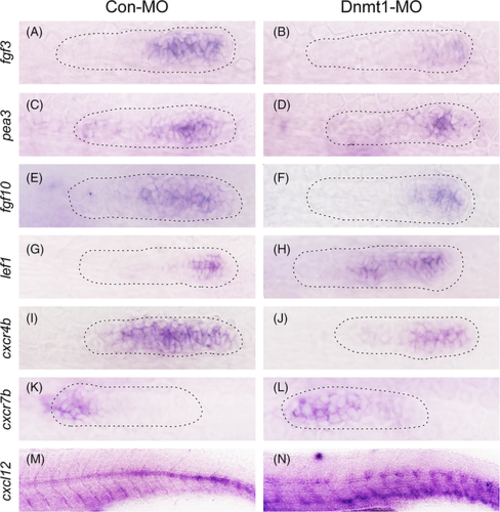
Dnmt1 depletion represses Fgf signalling and the chemokine superfamily in zebrafish primordium. Corresponding decreased expression of Fgf signalling components including fgf3, pea3 and fgf10 are presented in Dnmt1 morphants compared to the control siblings at 32 hpf (A-F). On the contrary, the expression of lef1, a target gene of Wnt signalling, is increased in Dnmt1-deficient embryos (G-H). The expression levels of chemokine ligands are discrepant that cxcr4b transcript level is decreased (I-J) but cxcr7b transcript level is increased (K-L) in Dnmt1-MO morphants in comparison with Con-MO samples. The staining of cxcl12 is completely discontinuous along the intermediate line of the trunk in Dnmt1-MO group compared to the controls (M-N). The primordium appearance is shaped by dotted lines. Each group has eight zebrafish, and the results are repeated for three times
|
|
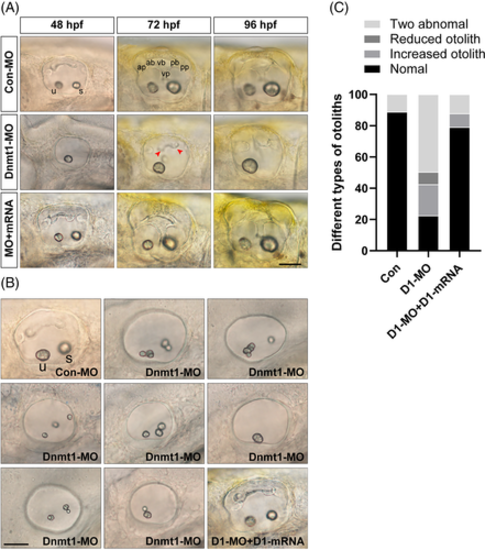
Knockdown of Dnmt1 disrupts the normal development of semicircular canals and otolith organs. A, Knockdown of Dnmt1 fails to form normal semicircular canals as well as otolith organs from 48 hpf to 96 hpf. Anterior, posterior and lateral (ventral) semicircular canals together with two otolith organs are well-structured in controls and in the mRNA rescue larvae, while in Dnmt1-MO morphants the normal semicircular canals are absent and the malformation of otolith maintain from 48 to 96 hpf. B, The inner ear structure of zebrafish at 48 hpf in white light in controls shows two normal otolith organs labelled as the anterior utricle and the posterior saccule, and upsides epithelial thickening. Multiple abnormal otolith phenotypes after Dnmt1 depletion are found at 48 hpf: increased number malformation, decreased number malformation and two abnormal otoliths. Successful rescue of phenotypic abnormalities is found in Dnmt1-MO + Dnmt1 mRNA groups. C, Different types of deformities and their percentages are compared among the three groups (n = 62, 420 and 81 in controls, Dnmt1-MO morphants and rescue embryos respectively). u: utricle, s: saccule, ap: anterior protrusion, ab: anterior bulge, vb: ventral bulge, vp: ventral protrusion, pb: posterior bulge and pp: posterior protrusion. Red arrow heads show the abnormal fusion of anterior and posterior protrusions. All images are lateral views with the anterior to the left and the dorsal side up. Scale bars are 50 μm (A, B)
|
|
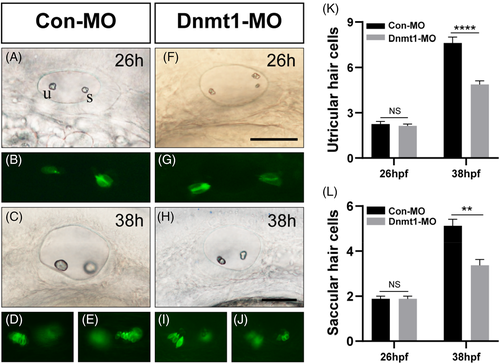
The HC differentiation both in utricle and saccule are interfered by Dnmt1 inhibition. At 26 hpf, the number of progenitor HCs are comparable in Con-MO and Dnmt1-MO group (A-B, F-G, K). At 38 hpf, reduced numbers of HCs are observed in Dnmt1-MO defects compared to the controls (C-D, H-J, L). Scale bars are 50 μm (A, C, F and H). Data are recorded as mean ± SEM. **p < 0.01, ****p < 0.0001
|
|
Changes in the expression of otic placode marker genes after Dnmt1 knockdown. In situ staining of pax2 and pax5 are all downregulated in Dnmt1-MO embryos compared to the controls by WISH at 48 hpf. The black dotted lines outline the otic vesicle. The Fgf signalling ligands fgf8, fgf3 and fgf10 are in lower expression in Dnmt1-MO embryos compared to the controls at 48 hpf
|
|
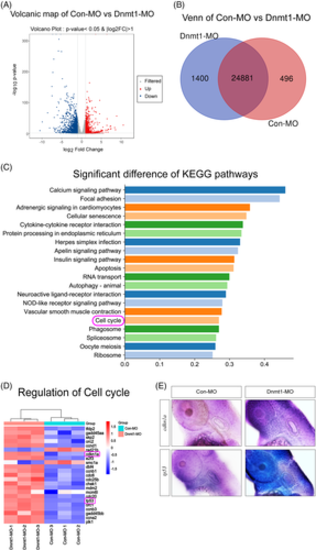
RNA-Seq analysis uncovers cell cycle genes as the key regulator of Dnmt1 inhibition during otic development of zebrafish. A, Volcanic plot analysis displays all the DEGs in the two groups. B, Venn map further labels the unique and overlapping number of DEGs in Con-MO and Dnmt1-MO groups. C, KEGG enrichment analysis exhibits the top 20 signalling pathways statistically different between Dnmt1 knockdown samples and the controls. D, Cluster analysis of DEGs on cell cycle pathway between the two groups. E, The ISH data confirm both cdkn1a (p21) and tp53 upregulated after knockdown of Dnmt1 compared to controls at 48 hpf
|
|
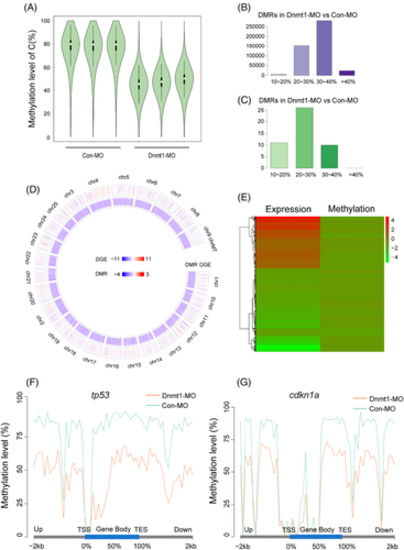
WGBS analysis detects global hypomethylation after knocking down Dnmt1. A, The violin diagrams show global methylation levels in Con-MO group and Dnmt1-MO group with three repeats. The white dot indicates median, the black solid frame indicates interquartile range (IQR), the thin black line indicates data range and the width of violin indicates the distribution density. B-C, The number of DMRs with hypomethylation (B) and hypermethylation (C) in Dnmt1-MO group compared to Con-MO group. The abscissa represents different methylation levels. D, Circos figure displays the general distribution of DEGs and DMRs in genome Dnmt1-MO group compared to Con-MO group. The chromosome number, the DEGs (RNA-Seq) and DMRs (CpG) are present from the outer ring to the inner ring. Red indicates upregulation, blue indicates downregulation; the darker the colour, the greater the difference. E, The heat map shows the common DEG- and DMR-related genes in Dnmt1-MO group compared to Con-MO group. Red indicates upregulation and green indicates downregulation. F-G, The methylation levels of tp53 and cdkn1a in the functional region are all significantly reduced in Dnmt1-MO morphants compared to the Con-MOs. TES, transcription end site; TSS, transcription start site
|