- Title
-
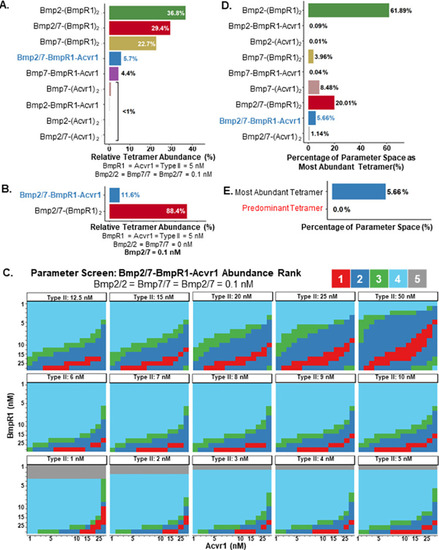
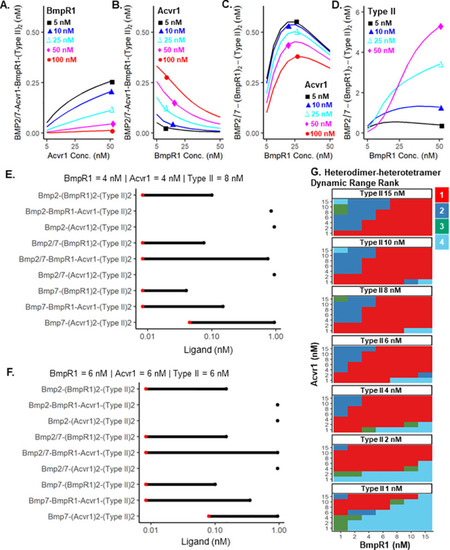
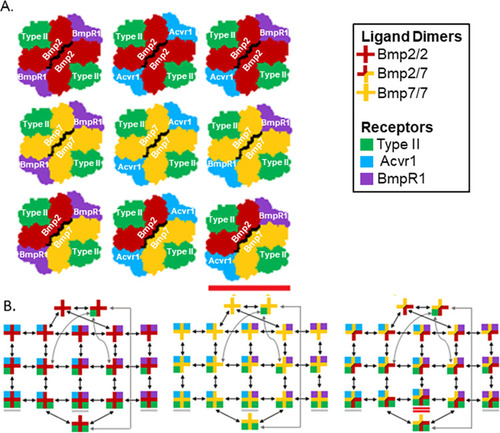
Heterodimer-heterotetramer formation mediates enhanced sensor activity in a biophysical model for BMP signaling
- Authors
- Karim, M.S., Madamanchi, A., Dutko, J.A., Mullins, M.C., Umulis, D.M.
- Source
- Full text @ PLoS Comput. Biol.
|
|
|
|
|
|
|
|