- Title
-
A two-site flexible clamp mechanism for RET-GDNF-GFRα1 assembly reveals both conformational adaptation and strict geometric spacing
- Authors
- Adams, S.E., Purkiss, A.G., Knowles, P.P., Nans, A., Briggs, D.C., Borg, A., Earl, C.P., Goodman, K.M., Nawrotek, A., Borg, A.J., McIntosh, P.B., Houghton, F.M., Kjær, S., McDonald, N.Q.
- Source
- Full text @ Structure
|
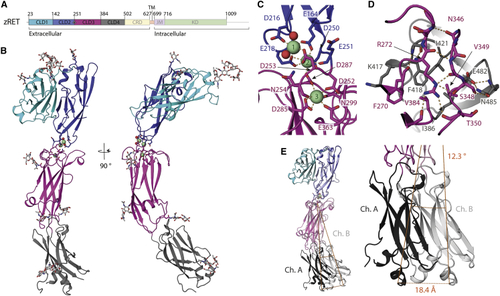
Crystal structure and flexibility of the zRET CLD(1-4) module (A) Schematic of zebrafish RET receptor tyrosine kinase. CLD, cadherin-like domains; CRD, cysteine-rich domain; TM, transmembrane helix; JM, juxtamembrane domain; KD, kinase domain. (B) Orthogonal views of zRETCLD1-4 colored as in (A). The calcium-binding site between CLD(2-3) has three calcium ions as green spheres with coordinating ligands as sticks and waters as red spheres. (C) Close-up view of the coordination shell for the three calcium atoms between CLD2 and CLD3. (D) Close-up of the interface between CLD3 and CLD4 centered on R272, selected side chains shown as sticks and dashed lines for hydrogen bonds. (E) Superposition of chains A and B within the crystallographic asymmetric unit. |
|
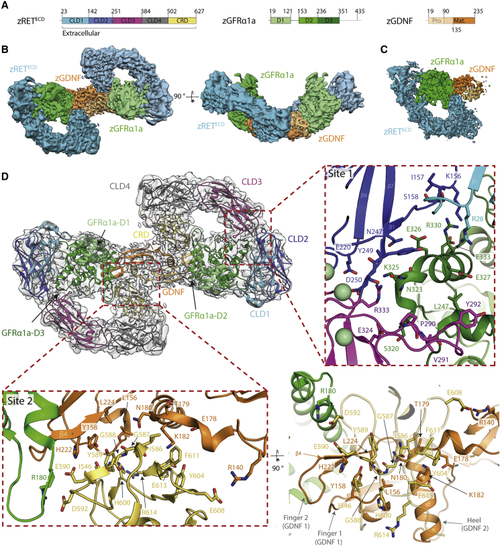
Cryo-EM structure of the zRETECD-zGFRα1aD1-3-zGDNFmat. (zRGα1a) complex (A) Schematic of zRETECD, zGFRα1aD1-3, and zGDNFmat., color coded according to (B) Orthogonal views of the reconstituted zRGα1a complex cryo-EM map, projecting down the approximate molecular dyad or perpendicular to it. The cryo-EM map is segmented and colored by protein, with zRETECD cyan, zGFRα1aD1-3 green, and zGDNFmat. orange. (C) Symmetry-expanded map of zRGα1a half-complex, with the map segmented and colored by protein as in (B). (D) The final model of the zRGα1a complex built into the C2 symmetry map, colored light gray. The domains are colored as in |
|
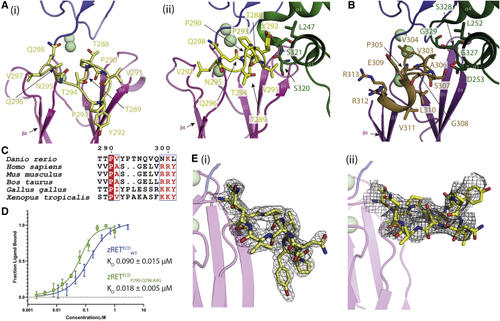
Ligand-co-receptor-induced conformational changes in zRETECD (A) The CLD3-β2-β3 loop is shown in yellow as sticks (i) projects “downward” in the view shown for zRET CLD(1-4) (see the orientation of Y292 side chain) and (ii) projects “upward” to engage the GFRα1D2 α1 helix (green sticks) in the zRGα1a structure. (B) The shorter CLD3-β2-β3 loop and extra helix from the human RETECD-NRTN-GFRα2 structure (PDB: (C) Sequence alignment of RET CLD3-β2-β3 loop sequences by Espript ( (D) Binding curves and calculated KD values for zRETECDwt and mutant (zRETECDP291-Q296:AAG) binding to zGFRα1a2-zGDNF2 measured by MST, with a minimum of n = 3 repeats for the WT and the mutations with the SE represented. (E) (i) Electron density map calculated using m2Fo-DFc coefficients over the CLD3-β2-β3 loop, yellow sticks and contoured at 1.0σ. (ii) Coulombic potential cryo-EM map for CLD3-β2-β3 loop from the zRGα1a complex (black mesh). Calcium ions are represented as pale green spheres. |
|
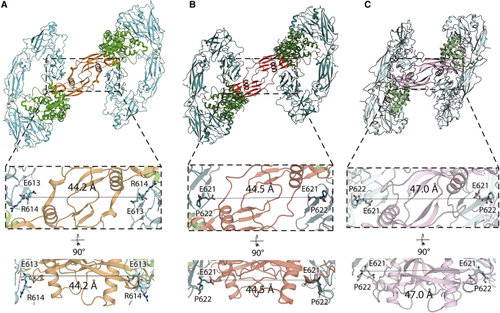
Different GFL ligands establish a conserved spacing between RET CRD-CRD pairs in their respective ternary complexes (A) Separation between the Cα of E613 (equivalent to E620 of hRET) from both molecules of zRETECD within the zRGα1a structure. (B) Equivalent distance between the Cα E620 from both molecules of hRETECD from the hRETECD-NRTN-GFRα2 (PDB: (C) Equivalent separation between the Cα E620 from the two molecules of hRETECD from the hRETECD-GDF15-GFRAL (PDB: |
|
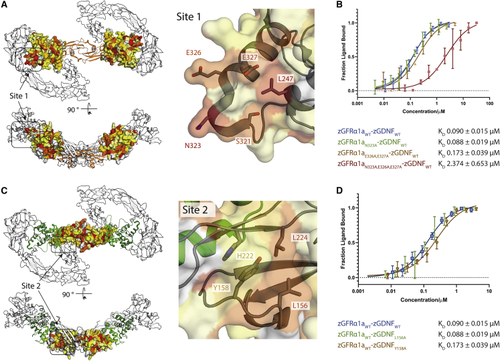
Mutational analysis of zGDNF and zGFRα1 site 1 and 2 interactions with zRETECD (A) Heatmap of the sequence conservation between hGFRα paralogs, and zGFRα1a mapped onto the structure of zGFRα1a D2-D3 domains reported here. Residues are colored by similarity (red highly similar to yellow through to white, least similar). Two orthogonal views are shown. Right panel, close-up of site 1 and conserved zGFRα1a residues. (B) Binding curves and KD values obtained using MST for zGFRα1aD1-3 and mutations assessed in complex with zGDNFmat., with a minimum of n = 3 repeats for the WT and the mutations with the SE represented. (C) Heatmap of the sequence similarity between GDNF paralogs depicted as a surface representation, mapped onto zGDNF138-235. Right panel, close-up of site 2 contact between RETCRD and zGDNF dimer. (D) MST binding curves and KD values for zGDNF and mutations L156A and Y158A probed in complex with WT zGFRα1a binding to zRETECD. |
|
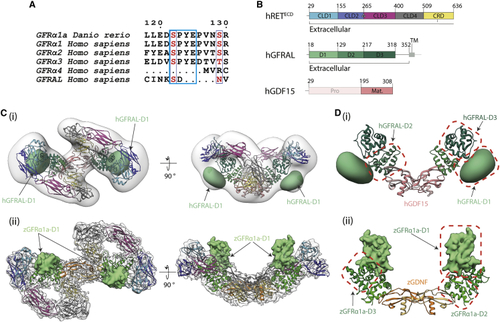
Divergent GFRα1/GFRAL co-receptor D1 domain positions within the RETECD ternary complex (A) The D1-D2 domain linker motif (SPYE), highlighted in cyan is conserved between zGFRα1a, GFRα1, GFRα2, and GFRα3. It is missing from the shorter GFRα4 that lacks a D1 domain altogether and from the divergent GFRAL. (B) Schematic diagram of human RETECD, GFRAL, and GDF15 construct boundaries used and individual domains annotated as in (C) (i) Negative stain EM envelope of a reconstituted hRETECD2-hGDF152-hGFRAL2 (hR15AL) complex docked with hR15AL (PDB: (D) Comparison of co-receptor D1 domain position and interfaces (i) GFRALD1 makes different contacts to domains D2-D3 (green), GFRALD1 shown as a 30 Å2 Gaussian volume (light green), and GDF15 (salmon). (ii) zGFRα1aD1 contacts and colored as in |
|
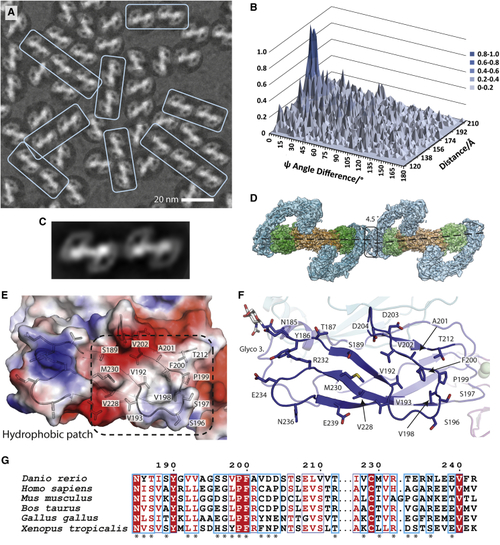
Evidence for linear arrays of zRGα1a particles on cryo-EM grids (A) Close-up of a representative micrograph for zRGα1a showing the particle orientation bias by fitting the dominant 2D class average view into picked particles and a recurring linear array of particles highlighted within pale cyan boxes. (B) Statistical distribution of the difference between the angle psi (Δψ) between two particles and their separation distance. Here the angle ψ is defined for each particle as the angle of rotation of each particle required to align it onto the 2D class average. (C) 2D class average from automated particle picking containing two adjacent zRGα1a particles. (D) The zRGα1a-zRGα1a interface highlighted with a black box. The angle and separation between each complex is based on the peak maxima coordinates from (B) assuming both particles are at the same Z height. (E) An electrostatic potential surface with selected side chains for the homotypic zRETCLD2 interface. (F) Close-up of the CLD2 contact, highlighting interface residues. (G) Conservation of representative RET sequences at the CLD2-CLD2 interface shown with an asterix. |
Reprinted from Structure (London, England : 1993), 29(7), Adams, S.E., Purkiss, A.G., Knowles, P.P., Nans, A., Briggs, D.C., Borg, A., Earl, C.P., Goodman, K.M., Nawrotek, A., Borg, A.J., McIntosh, P.B., Houghton, F.M., Kjær, S., McDonald, N.Q., A two-site flexible clamp mechanism for RET-GDNF-GFRα1 assembly reveals both conformational adaptation and strict geometric spacing, 694-708.e7, Copyright (2021) with permission from Elsevier. Full text @ Structure