Figure 5
Mutational analysis of zGDNF and zGFRα1 site 1 and 2 interactions with zRETECD
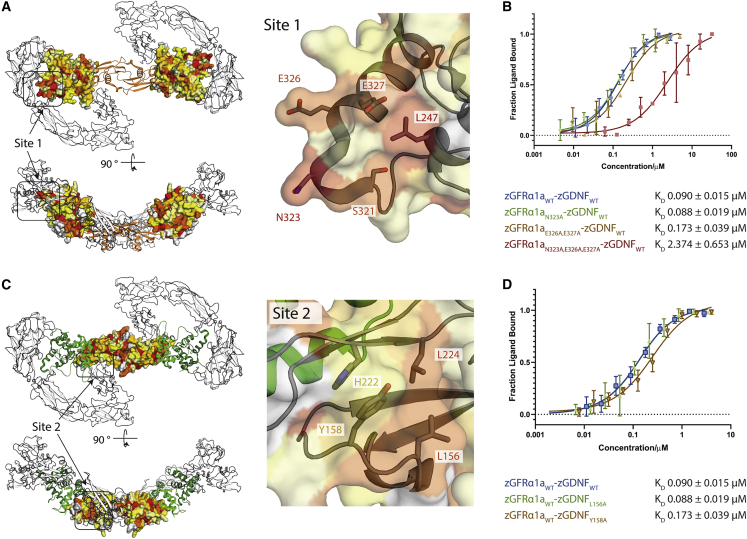
(A) Heatmap of the sequence conservation between hGFRα paralogs, and zGFRα1a mapped onto the structure of zGFRα1a D2-D3 domains reported here. Residues are colored by similarity (red highly similar to yellow through to white, least similar). Two orthogonal views are shown. Right panel, close-up of site 1 and conserved zGFRα1a residues.
(B) Binding curves and KD values obtained using MST for zGFRα1aD1-3 and mutations assessed in complex with zGDNFmat., with a minimum of n = 3 repeats for the WT and the mutations with the SE represented.
(C) Heatmap of the sequence similarity between GDNF paralogs depicted as a surface representation, mapped onto zGDNF138-235. Right panel, close-up of site 2 contact between RETCRD and zGDNF dimer.
(D) MST binding curves and KD values for zGDNF and mutations L156A and Y158A probed in complex with WT zGFRα1a binding to zRETECD.
Reprinted from Structure (London, England : 1993), 29(7), Adams, S.E., Purkiss, A.G., Knowles, P.P., Nans, A., Briggs, D.C., Borg, A., Earl, C.P., Goodman, K.M., Nawrotek, A., Borg, A.J., McIntosh, P.B., Houghton, F.M., Kjær, S., McDonald, N.Q., A two-site flexible clamp mechanism for RET-GDNF-GFRα1 assembly reveals both conformational adaptation and strict geometric spacing, 694-708.e7, Copyright (2021) with permission from Elsevier. Full text @ Structure