- Title
-
An image-based data-driven analysis of cellular architecture in a developing tissue
- Authors
- Hartmann, J., Wong, M., Gallo, E., Gilmour, D.
- Source
- Full text @ Elife
|
( |
|
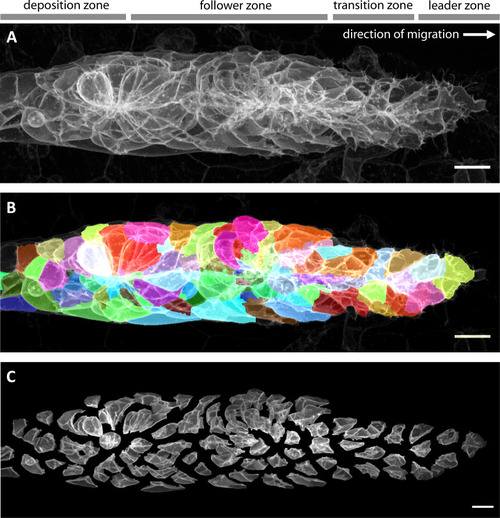
(A) Maximum z-projection of a deconvolved 3D volume of the pLLP acquired using the LSM880 AiryScan FAST mode. (B) The same primordium shown with a semi-transparent color overlay of the corresponding single-cell segmentation. (C) Expanded view of the same primordium; individual segmented cells have been shifted apart without being rescaled or deformed, revealing their individual shapes within the collective. Note that the segmentation faithfully recapitulates the diversity of cell shapes within the pLLP, with the exception of fine protrusions. Since the protrusions of follower cells are often impossible to detect against the membranes of the cells ahead of them, we decided not to include fine protrusions in our analysis. All scale bars: 10 μm. |
|
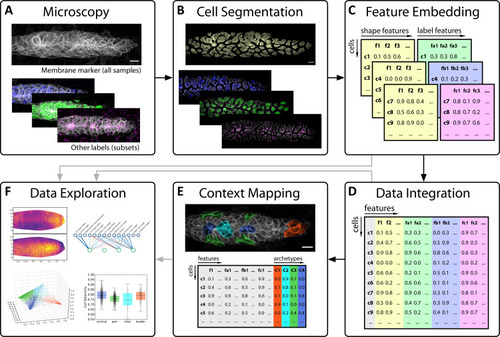
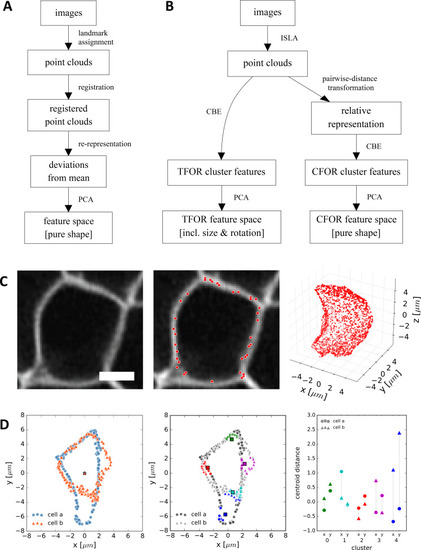
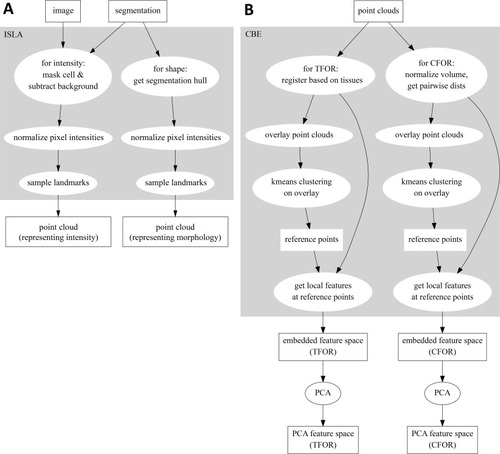
(A) A classical workflow in landmark-based geometric morphometrics. (B) Adapted workflow for morphometrics of arbitrary fluorescence intensity distributions. See Figure 3—figure supplement 1 for a more detailed version. (C) Illustration of ISLA, our algorithm for conversion of voxel-based 3D images to representative point clouds. Shown are a slice of an input image (left), here a membrane-labeled cell in the pLLP (scale bar: 2 μm), the landmarks sampled from this image (middle), here oversampled compared to the standard pipeline for illustration purposes, and the resulting 3D point cloud (right). (D) Illustration of CBE, our algorithm for embedding point clouds into a feature space. In this 2D mock example, two cells are being embedded based on point clouds of their outlines (left). CBE proceeds by performing clustering on both clouds combined (middle) and then extracting the distances along each axis from each cluster center to the centroid of its ten nearest neighbors (right). Note that the most distinguishing morphological feature of the two example cells, namely the outcropping of cell a at the bottom, is reflected in a large difference in the corresponding cluster's distance values (cluster 4, blue). |
|
( |
|
( |
|
( |
|
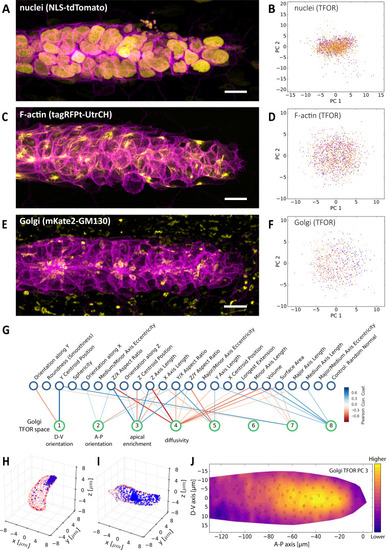
(A, C, E) Maximum z-projections of two-color stacks showing the membrane in magenta and one of three subcellular structures in yellow. (B, D, F) Tissue frame of reference (TFOR) CBE embeddings corresponding to the three structures shown in A, C and E. The different colors of points indicate different primordia. The three structures are nuclei (N = 20, n = 2528) (A–B), F-actin (N = 19, n = 1876) (C–D) and the Golgi apparatus (N = 11, n = 866) (E–F). (G) Bigraph showing correlations between the Golgi's embedded features and our engineered cells shape features (see Supplementary file 2). The first two Golgi TFOR PCs match those found in the cell shape TFOR space (see Figure 4E) whereas PCs 3 and 4 are specific to the Golgi. For technical details see the legend of Figure 4E. (H–I) Point cloud renderings showing the distribution of Golgi signal (blue, membranes in red) in two example cells, one with a high value in the Golgi's TFOR PC 3 (H) and one with a low value (I), illustrating that PC three captures apical enrichment of the Golgi. (J) Consensus tissue map for Golgi PC 3 (apical enrichment), showing increased values behind the leader zone. For technical details see the legend of Figure 4G. |
|
( |
|
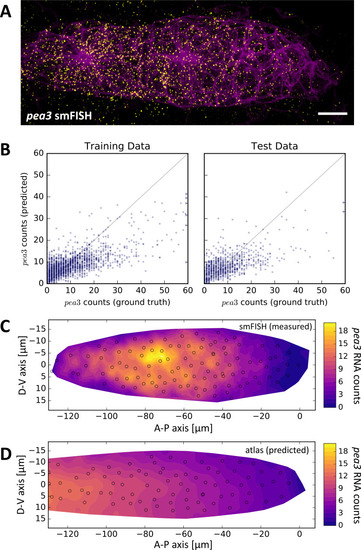
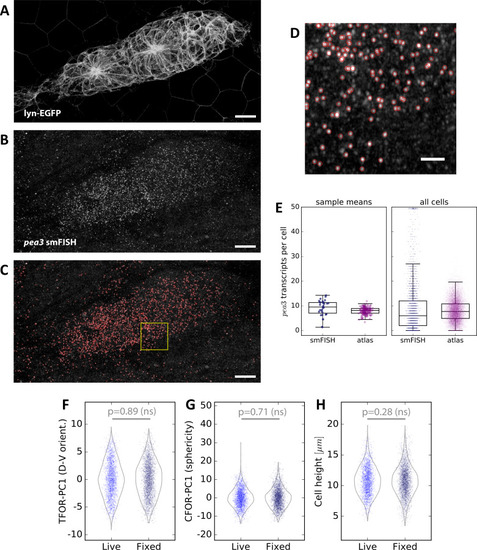
(A) Maximum z-projection of a two-color stack of pea3 smFISH (yellow) and the lyn-EGFP membrane marker (magenta). Scale bar: 10 µm. (B) Results of SVR regression on pea3 spot counts using TFOR and CFOR shape features as well as cell centroid coordinates of registered primordia as input. Each blue dot is a cell, the diagonal gray line reflects perfect prediction and blue arrows at the border point to outliers with very high spot counts. On training data, the regressor's explained variance ratio is 0.462 ± 0.011, on previously unseen test data it achieves 0.382 ± 0.019. (C–D) Consensus tissue maps of pea3 expression generated directly from the pea3 smFISH dataset (C) or from the full atlas dataset based on SVR predictions of spot counts (D). Note that the prediction for the entire atlas preserves the most prominent pattern – the front-rear gradient across the tissue – but does not capture the noisy heterogeneity among follower cells observed in direct measurements. |
|
( |
|
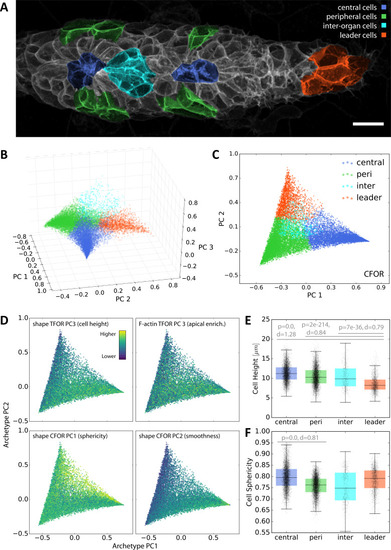
(A) A maximum z-projected example stack with colors highlighting different conceptual archetypes in the pLLP that have been manually annotated. (B) A low-dimensional archetype space resulting from a PCA of the SVC prediction probabilities (with the SVC having been trained on CFOR shape features). Cells are placed according to how similar they are to each archetype, with those at the corners of the tetrahedron belonging strictly to the corresponding archetype and those in between exhibiting an intermediate morphology. (C) Since inter-organ cells are not morphologically distinct enough at this stage (see Figure 7—figure supplement 1), the archetype space can be reduced to 2D without much loss of information. (D) Scatter plots of the 2D archetype space with additional information from the cellular shape space and from the protein distribution atlas superimposed in color. (E–F) Boxplots showing data grouped by predicted archetype labels. This form of grouping allows statistical analysis, showing that leader cells are flatter than any other class of follower cells (E) and that central rosette cells are more spherical than peripheral rosette cells (F). Whiskers are 5th/95th percentiles, p-values are computed with a two-sided Mann-Whitney U-test, and Cohen's d is given as an estimate of effect size. |
|
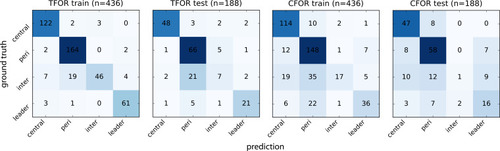
Confusion matrices for SVC archetype classification. The ground truth is based on manual annotation of high-confidence cases. Note that using TFOR features results in slightly better performance than using CFOR features, implying that rotational information and cell size are useful for prediction to some extent. Overall prediction accuracy is high but inter-organ cells are frequently mislabeled, in particular as peripheral cells, indicating that they are morphologically similar at this stage and thus difficult to distinguish based on shape features alone. |