- Title
-
Nkx6 proteins specify one zebrafish primary motoneuron subtype by regulating late islet1 expression
- Authors
- Hutchinson, S.A., Cheesman, S.E., Hale, L.A., Boone, J.Q., and Eisen, J.S.
- Source
- Full text @ Development
|
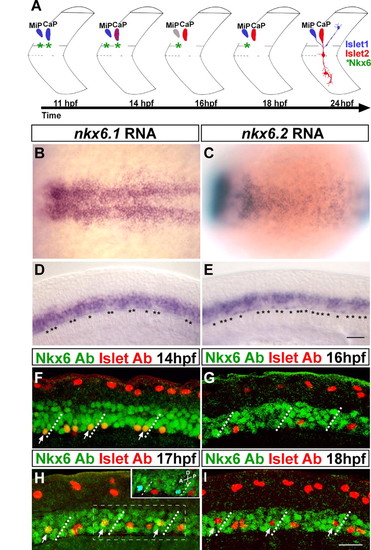
Two Nkx6 proteins are dynamically expressed in PMNs. (A) Schematic of Islet and Nkx6 protein dynamics during PMN subtype specification at the 8- to 12-somite level. At 11 hpf, MiP and CaP both express Islet1 (blue) and Nkx6 (green asterisks). At 14 hpf, CaP and MiP maintain Nkx6 and Islet1 expression and CaP has initiated expression of Islet2 (red). At 16 hpf, Islet1 is downregulated in MiP (gray); CaP has downregulated expression of Nkx6 and Islet1, but continues to express Islet2. By 18 hpf, MiP has reinitiated expression of Islet1. At 24 hpf, neither MiP nor CaP expresses Nkx6; MiP still expresses Islet1, and CaP still expresses Islet2. (B,C) Dorsal views of 10-hpf embryos. (B) nkx6.1 mRNA is expressed in many medial neurectodermal cells in the pMN domain. (C) nkx6.2 mRNA is expressed in the same domain as nkx6.1. (D,E) Lateral views of 18-hpf embryos showing that (D) nkx6.1 and (E) nkx6.2 mRNA is expressed in the ventral spinal cord; however, some of the ventral-most cells are nkx6-negative (asterisks). (F-I) Lateral views of Islet and Nkx6 co-expression. MiP and CaP were identified by soma position relative to somite boundaries (dotted lines). (F) Single optical slice showing that all ventrally located PMNs express Islet and Nkx6 proteins at 14 hpf, whereas dorsally located Rohon-Beard neurons express only Islet. (G-I) Projected z-stacks showing localization of Nkx6 and Islet Abs in the ventral neural tube. (G) By 16 hpf, CaPs no longer express Nkx6. (H) At 17 hpf, MiPs (arrows) express Nkx6, whereas CaPs are Nkx6-negative. Inset is a 3D rendering of the boxed area; cells co-expressing Nkx6 and Islet (arrows in H) were identified by the Velocity software program (see Materials and methods) and are pseudo-colored blue. The slight differences between the boxed area in H and the inset are due to the differences in angle of visualization: H is a direct lateral view, whereas the inset is rotated (see axes in inset). (I) By 19 hpf, MiPs (arrows) have also downregulated Nkx6. Scale bars: 50 µm in B-E; 20 µm in F-I. EXPRESSION / LABELING:
|
|
Nkx6 proteins regulate late islet1 expression. (A-D) Dorsal views at 11-12 hpf. (A) islet1 mRNA expression in PMNs of control embryos. islet1 mRNA expression is unaffected by nkx6.1 (B), nkx6.2 (C) or double nkx6 (D) MO-injection. (E-L) Lateral views of axial level 8-12 at 18 hpf. (E-H) islet2 mRNA expression is normal in control and nkx6 MO-injected embryos. (I-L) islet1 mRNA expression is confined to MiPs (arrows). In nkx6.1 MO-injected embryos, there are fewer islet1-positive cells (J; islet1-positive cells present in 74.4% of segments; n=133 segments in 14 embryos) as compared with controls (I; islet1-positive cells present in 82.4% of segments; n=97 segments in 12 embryos). The faint cell in J in the segment lacking an arrow is on the other side of the embryo; as far as we can tell the distribution of segments lacking islet1-positive MiPs is random and not correlated between the two sides of the embryo. (K) nkx6.2 MO-injected embryos also had a decrease in islet1 expression (islet1-positive cells present in 67.7% of segments; n=99 segments in 10 embryos). (L) The most dramatic loss of islet1 expression was in double nkx6 MO-injected embryos (islet1-positive cells present in 58.7% of segments; n=126 segments in 13 embryos). The number of islet1-positive cells might be an overestimate in some cases, because of islet1 RNA expression in RoP motoneurons (Appel et al., 1995). In some of these images, there appear to be abnormal numbers of Rohon-Beard (RB) spinal sensory neurons (islet1-positive cells in the dorsal spinal cord). This is an artifact of the way we mount the embryos to visualize PMNs in focus in many adjacent segments, which often requires tilting the embryos. The number of RBs appears within the normal range in nkx6 MO-injected embryos. Scale bar: 20 µm. EXPRESSION / LABELING:
|
|
MiP axons are absent from embryos lacking Nkx6 proteins. (A-D) Lateral views at axial level 8-12 of 28-hpf embryos labeled with zn1 and znp1 Abs. Arrowheads point to CaP axons; arrows point to MiP axons; dashed lines indicate MiP axon trajectories. Embryos lacking Nkx6.2 or Nkx6.1 (B,C) have normal CaP axons, but MiP axons were absent, truncated or excessively branched compared with control embryos (A). (D) Loss of MiP axons is most severe in double nkx6 MO-injected embryos, but CaP axons are normal. (E) Loss of MiP axons in embryos lacking Nkx6.1 and Nkx6.2 is restored by co-injection of islet1 mRNA with the nkx6 MOs. (F) Quantification of segments lacking MiP axons in control, nkx6 MO-injected, and nkx6 MO plus islet1 mRNA-injected embryos. Scale bar: 20 µm. PHENOTYPE:
|
|
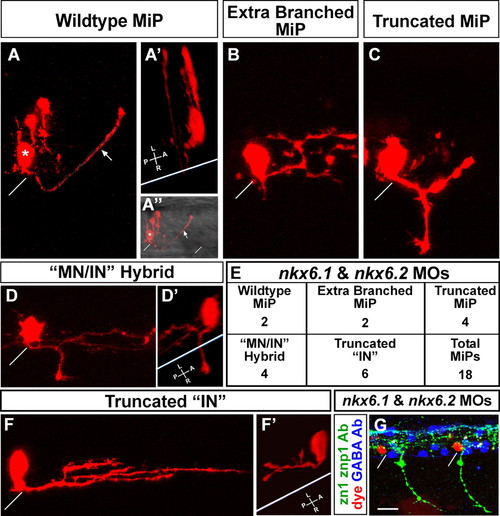
MiPs become more interneuron-like in the absence of Nkx6 proteins. (A-F′) Single cells in the MiP position were identified and labeled at 24 hpf as described by Eisen et al. (Eisen et al., 1989); images were captured at 28 hpf. (A) Wild-type MiP, with its characteristic dorsal axon. The diagonal white line shows the location of the overlying segment boundary (likewise in A″,B,C,D,F,G). The asterisk shows the MiP cell body. The arrows (A,A″) indicate the MiP dorsal axon. Two other cells were also labeled during micropipette penetration of the spinal cord; these are located just dorsal of the MiP cell body. (A′) 3D rotation of the confocal image. The white line indicates the ventral aspect of the spinal cord (likewise in D′ and F′). Note that the MiP dorsal axon loops around this line, showing that it extends out of the spinal cord. (A″) The same view as A, but the fluorescent image is merged with a brightfield image to show the ventral aspect of the spinal cord and the overlying segment boundaries. (B-E) Range of phenotypes of MiPs in embryos lacking Nkx6 proteins. (B) This cell has a normal ventral axon remnant, but the dorsal axon is truncated and excessively branched. (C) This cell has abnormally retained the ventral axon and the dorsal axon is truncated. (D) This cell has abnormally retained the ventral axon; the 3D rotation in D′ shows that it extends out of the spinal cord. The cell has not developed a dorsal axon, but instead has an interneuron-like axon within the spinal cord, as shown in the rotation in D′. (E) Quantification of phenotypes shown in B-D and F. IN, interneuron; MN, motoneuron. (F,F′) This cell has neither a ventral nor a dorsal axon, but has an excessively branched interneuron-like axon that is truncated relative to those of wild-type interneurons. The 3D rotation in F′ shows that this interneuron-like axon is entirely within the spinal cord. (G) Triple label showing that dye-labeled MiPs (red) are located in the normal MiP position, are adjacent to GABA-positive interneurons (blue) in the VeLD, KA′ and KA″ positions (white asterisks above cells in the VeLD position), and are in segments with normal CaP axons but lacking MiP axons as revealed by labeling with zn1 and znp1 Abs (green). Scale bar: 10 μm in A-D′,F,F&prime& 20 μm in G. PHENOTYPE:
|
|
Nkx6 proteins regulate eng1b and chx10 expression. (A-F) Lateral views of segments 8-12. (A,B) 28-hpf embryos labeled with GABA Ab. GABA-positive cells in the VeLD and KA′ positions (denoted V-K) (see Hutchinson and Eisen, 2006) are labeled with white asterisks. nkx6 MO-injected embryos (B) have a similar number of ventral GABA-positive interneurons as control embryos (A). (C,D) 24-hpf embryos labeled with chx10 riboprobe. nkx6 double MO-injected embryos (D) have fewer chx10-positive cells than control embryos (C). (E,F) 24-hpf embryos labeled with eng1b riboprobe. nkx6 double MO-injected embryos (F) have more eng1b- positive cells than control embryos (E). (G) Quantification of GABA-positive, chx10- positive, and eng1b-positive cells in control and nkx6 double MO-injected embryos. Scale bars: 20 μm in A,B; 50 μm in C-F. EXPRESSION / LABELING:
|
|
Zebrafish have three Nkx6 proteins. (A) Nkx phylogeny demonstrating the three zebrafish Nkx6 proteins uniquely cluster within one node which is distinct from the Nkx2.2 and Nkx1 class of proteins. The PHYLIP software package was employed to create a neighbor-joining tree using the ClustalW algorithm. Sequences included in the tree were gathered from GenBank and bootstrap values are indicated. Expression of nkx6.3 mRNA begins at 20 hpf, persists until at least 72 hpf, and is limited to a small population of cells posterior to rhombomere 5 in the hindbrain. (B) Alignment of zebrafish Nkx6.1, Nkx6.2 and Nkx6.3 translated amino acid sequences. The dashed red line indicates the NK domain and the solid blue line indicates the homeodomain. (C) Whole-mount in situ RNA hybridization reveals that nkx6.3 is expressed in a subset of hindbrain cells at 25 hpf. (D) At 36 hpf, nkx6.3-expressing cells (purple) are Islet:GFP-negative and are located ventral to cells labeled with Islet:GFP (brown, GFP antibody staining). Lateral views, anterior to the left, dorsal to the top. ov, otic vesicle; sb, somite boundary. Scale bar: 40 μm. |
|
Anti-Nkx6 detects both Nkx6.1 and Nkx6.2 proteins. 24-hpf embryos, anterior to the left. (A-D,H-J) lateral views; (E-G) dorsal views. The confocal images in E-G were all taken at the same settings to ensure comparability; likewise for the confocal images in H-J. The confocal images in H-J show only one side of the spinal cord; thus, there are fewer labeled cells than in the images in A and B, which show whole-mount embryos. nkx6.1 (A) and nkx6.2 (B) mRNA are expressed in the ventral spinal cord in similar patterns. Both nkx6.1 and nkx6.2 (C,D) mRNA are expressed in the ventral brain, although the expression patterns are different. Injection of nkx6.1 MO decreased Nkx6.1 antibody labeling in brain (F) and spinal cord (I) as compared to controls (E,H). However, some Nkx6.1-positive cells remained, suggesting that the antibody recognizes both Nkx6.1 and Nkx6.2 proteins. We tested this hypothesis by co-injecting nkx6.1 and nkx6.2 MOs, which extinguished nearly all Nkx6 antibody labeling in the spinal cord (J) and brain (G). Thus, we infer that the antibody recognizes both Nkx6.1 and Nkx6.2 proteins. Scale bars: 50 μm. |

Unillustrated author statements EXPRESSION / LABELING:
|