- Title
-
A Dynamic Epicardial Injury Response Supports Progenitor Cell Activity during Zebrafish Heart Regeneration
- Authors
- Lepilina, A., Coon, A.N., Kikuchi, K., Holdway, J.E., Roberts, R.W., Burns, C.G., and Poss, K.D.
- Source
- Full text @ Cell
|
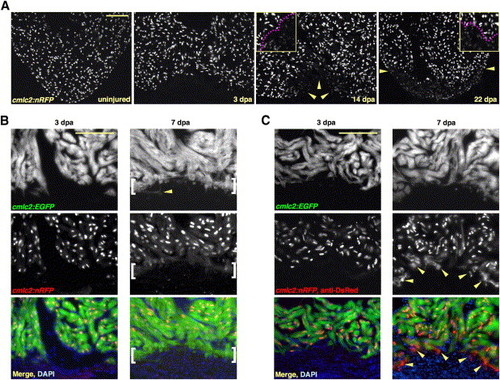
New Myocardium Arises from Undifferentiated Progenitor Cells (A) Sections through uninjured and injured cmlc2:nRFP ventricles. A subpopulation of RFPlo nuclei manifests by 14 dpa (arrowheads), representing ostensibly the entire regenerate by 22 dpa. Magenta line in high-magnification insets delineates RFPhi CMs (above line) from RFPlo CMs. (B) cmlc2:nRFP; cmlc2:EGFP ventricles. EGFP and RFP expression from the cmlc2 promoter is reported at the same basoapical level at 3 dpa. By 7 dpa, an RFPneg front of newly differentiated muscle reporting the faster-fluorescing EGFP (brackets) appears apical to the EGFPposRFPpos portion. Arrowhead indicates an EGFPpos cell process extending into the clot. (C) cmlc2:nRFP; cmlc2:EGFP ventricles stained for DsRed immunoreactivity with an anti-DsRed antibody. At 3 dpa, there is no difference in appearance from the unstained 3 dpa ventricle in (B). By 7 dpa, a front of RFPcyto muscle (arrowheads), representing the most recently differentiated CMs, colabels EGFPpos tissue apical to natural RFPnuc fluorescence. Scale bar = 100 μm. |
|
A Precardiac Field Is Established during Regeneration and Supports CM Proliferation (A) The precardiac marker hand2 is expressed in cells coating the apical edge of the existing muscle at 3 dpa (violet stains). At 4 dpa, nkx2.5 and tbx20 are also expressed in cells distributed along the resection plane. By contrast, no new CM differentiation events are detected by anti-DsRed staining in cmlc2:nRFP zebrafish at 3 dpa (see Figure 1) and only sparse events at 4 dpa (Table S1). The pink background represents myocardial autofluorescence, which aids visualization of positive signals. (B) nkx2.5, tbx20, hand2, tbx5, and Mef2 (red nuclear stain) are each present in a domain of enhanced expression at the apical edge of the regenerate (brackets), an area of most recent myocardial differentiation events as assessed in the cmlc2:nRFP; cmlc2:EGFP strain. The ISH pattern of cmlc2 at 7 dpa is also shown to indicate differentiated muscle. (C) By 14 dpa, freshly differentiated RFPcyto cells in cmlc2:nRFP; cmlc2:EGFP hearts appear smaller and remain at the apical edge as regeneration proceeds (brackets). Enhanced nkx2.5, tbx20, and hand2 expression remains detectable in this region. (D) Seven (left) and 14 dpa (right) ventricles from cmlc2:nRFP zebrafish stained for DsRed immunoreactivity (red) and BrdU incorporation (green). At 7 dpa, BrdU label is detectable only in newly differentiated RFPcyto cells (arrowheads). By 14 dpa, BrdU is also detectable in RFPnuc CMs (arrows). Scale bar = 100 μm. EXPRESSION / LABELING:
|
|
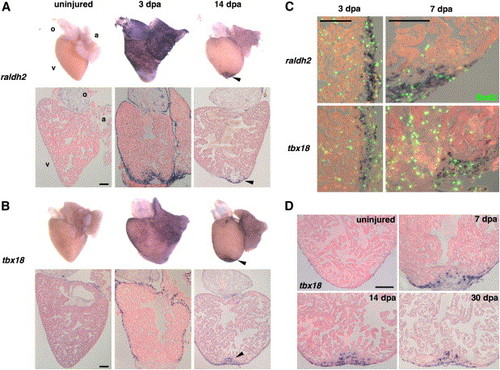
Organ-Wide Activation of Epicardial Cells and Invasion of the Regenerate (A and B) Whole-mount (top) and section (bottom) ISH of uninjured and 3 and 14 dpa hearts for raldh2 (A) and tbx18 (B). Expression of each marker is low or undetectable in the uninjured hearts but induced at 3 dpa in epicardial tissue surrounding the atrium (a) and ventricle (v) for tbx18 and these tissues plus the outflow tract (o) for raldh2. The endocardium surrounding wounded myofibers also expresses raldh2. By 14 dpa, expression of both markers is localized to the apical wound (arrowhead). (C) Colocalization of BrdU (green) with raldh2 (top) and tbx18 (bottom) in the ventricular epicardium at 3 and 7 dpa. The left images display a lateral edge of the 3 dpa ventricle away from the wound, while the right images display a lateral and apical portion of the 7 dpa ventricle including some of the wound. By 7 dpa, raldh2/tbx18/BrdU-positive cells begin to localize to the injury site. The majority of nonepicardial BrdU-positive cells are erythrocytes within the ventricular lumen. (D) Epicardial-derived cells positive for tbx18 invade the regenerate by 7 dpa and remain by 30 dpa. Scale bar = 100 μm. EXPRESSION / LABELING:
|
|
Expression of fgf17b, fgfr2, and fgfr4 during Heart Regeneration (A) Expression of fgf17b in the uninjured, 7, 14, and 30 dpa heart visualized by ISH. Weak, perinuclear expression was observed in uninjured ventricular CMs. Starting at 7 dpa, myofibers at the apical edge of the regenerate showed enhanced fgf17b expression (arrowheads), as did other cells within and around the injury. This augmentation persisted in regenerating muscle through 30 dpa. (B) Expression of fgfr2 during heart regeneration. While the uninjured ventricle showed no fgfr2 expression, epicardial cells induced fgfr2 upon entering the wound at 7 dpa (arrowheads), where expression was strongest at 14 dpa. By 30 dpa, faint expression was observed in isolated cells within the regenerate (arrowheads). (C) fgfr4 is expressed in a small number of epicardial or subepicardial cells on the uninjured ventricle (arrowheads) and induced in cells adjacent to regenerating muscle at 7 dpa and 14 dpa. fgfr4-expressing cells remain detectable in the regenerate by 30 dpa; in this image, fgfr4 expression in the valve can also be observed (upper right corner). Scale bar = 100 μm. EXPRESSION / LABELING:
|
|
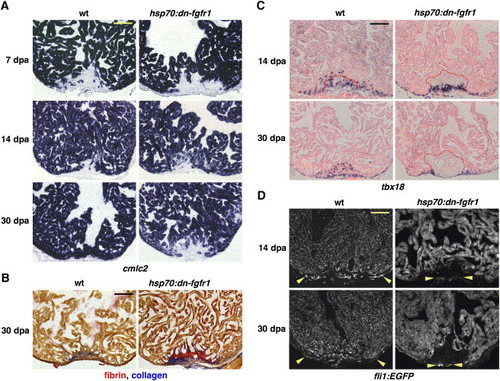
Fgfr Inhibition Blocks Epicardial EMT, Disrupting Coronary Neovascularization and Arresting Regeneration (A) cmlc2 expression at 7, 14, and 30 dpa in heat-induced wild-type and hsp70:dn-fgfr1 zebrafish. Transgenic animals arrested regeneration of cmlc2-positive differentiated muscle around 14 dpa, leaving a large wound by 30 dpa. (B) Thirty dpa hearts stained with acid fuchsin-orange G (AFOG). Wild-type ventricles display small deposits of scar tissue (blue) within a restored myocardial wall, while hsp70:dn-fgfr1 hearts retain large amounts of fibrin (orange) and collagen (blue). (C) Wild-type and hsp70:dn-fgfr1 ventricles at 14 and 30 dpa, stained for tbx18 expression by ISH. Wild-type tbx18-positive cells integrate into the regenerating muscle, but hsp70:dn-fgfr1 tbx18-positive cells fail to integrate and accumulate at the apical edge of the wound. Clot material is outlined in red. (D) Wild-type fli1:EGFP and hsp70:dn-fgfr1; fli1:EGFP ventricles at 14 and 30 dpa. Wild-type regenerates are well vascularized (region within arrowheads). By contrast, transgenic wounds developed little or no organized endothelial structures in the vicinity of muscle. Occasionally, vessels were seen on the periphery of the wound, as indicated by arrowheads in these ventricles. hsp70:dn-fgfr1 ventricles also display heat-induced fluorescence from the EGFP-tagged transgene. Scale bar = 100 μm. EXPRESSION / LABELING:
|
|
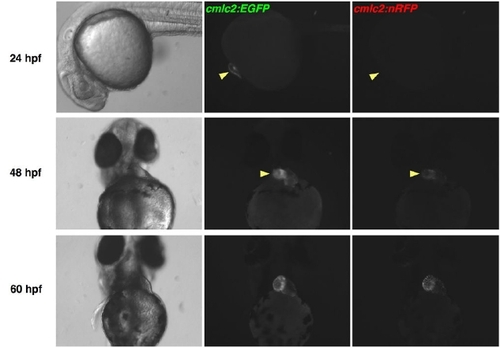
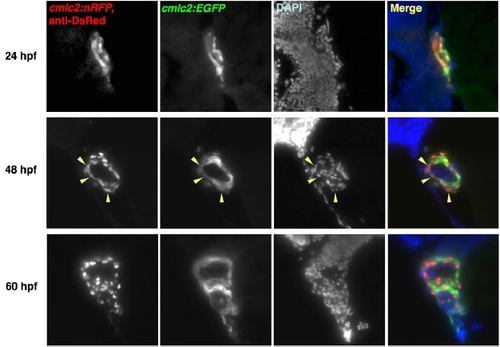
EGFP Matures More Rapidly than DsRed2 in Zebrafish Hearts. cmlc2:EGFP; cmlc2:nRFP double transgenic embryos were examined for fluorescence using a stereomicroscope. Cardiac EGFP fluorescence was easily identified at 24 hpf, while natural RFP fluorescence was first detectable by 48 hpf, approaching the intensity of EGFP fluorescence by 60 hpf. Fluorescent images have identical exposure times. |
|
DsRed Immunoreactivity in Freshly Differentiated Embryonic cmlc2:nRFP CMs Transitions from Cytosol to Nucleus. Histological sections of cmlc2:EGFP; cmlc2:nRFP double transgenic embryos were sectioned and stained for DsRed immunofluorescence. DsRed immunoreactivity mimicked natural cytosolic EGFP fluorescence at 24 hpf, was distributed in the cytosol of a minority of CMs at 48 hpf (arrowheads), and was nuclear at 60 hpf. |
|
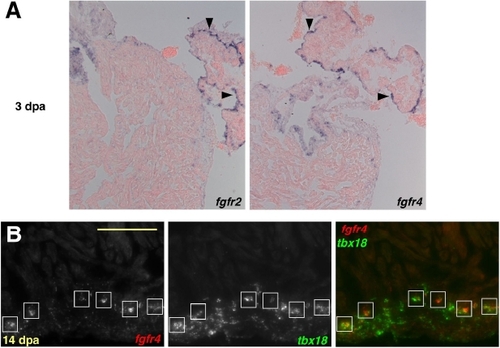
Epicardial Expression of fgfrs during Heart Regeneration. (A) At 3 dpa, fgfr2 and fgfr4 are expressed in contiguous domains of atrial epicardial cells (violet stains,arrowheads), distant from the injured ventricular apex (not shown). (B) Double in situ hybridization colocalizing fgfr4 (red) and tbx18 (green) at 14 dpa, demonstrating fgfr4 expression in epicardial-derived cells. fgfr4 was expressed in the tbx18-positive cells closest to the apical edge of the regenerate (boxes). Scale bar = 100 μm. EXPRESSION / LABELING:
|
|
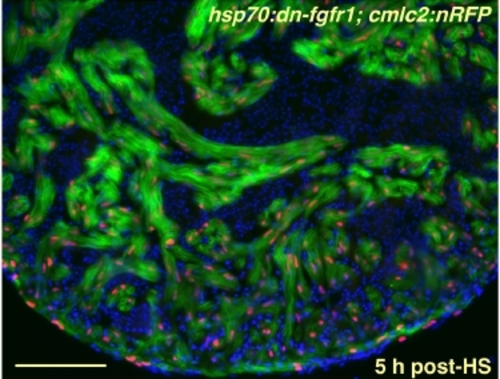
Strong, Uniform Induction of a dn-fgfr1 Transgene in the Uninjured Zebrafish Heart. Ventricular section from an uninjured hsp70:dn-fgfr1; cmlc2:nRFP double transgenic animal, 5 hours after heat-shock (HS). Cardiac cells, but not red blood cells, express EGFP for at least 24 hours post-shock. Red nuclei = CMs; green = heat-induced fluorescence; blue = nuclei. Scale bar = 100 μm. |
|
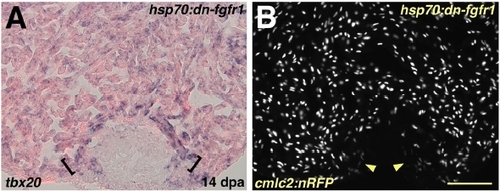
Myocardial Differentiation per se is Grossly Normal during Fgfr Inhibition (A) Induction of tbx20 in a hsp70:dn-fgfr1 ventricle at 14 dpa. A band of enhanced tbx20 expression at the distal edge of the regenerate adjacent to the wound is indicated by brackets. (B) Ventricular section from a hsp70:dn-fgfr1; cmlc2:nRFP double transgenic animal visualized for RFP fluorescence at 14 dpa. Arrowheads indicate RFPlo CMs. Scale bar = 100 μm. EXPRESSION / LABELING:
|
Reprinted from Cell, 127(3), Lepilina, A., Coon, A.N., Kikuchi, K., Holdway, J.E., Roberts, R.W., Burns, C.G., and Poss, K.D., A Dynamic Epicardial Injury Response Supports Progenitor Cell Activity during Zebrafish Heart Regeneration, 607-619, Copyright (2006) with permission from Elsevier. Full text @ Cell