- Title
-
The MITF paralog tfec is required in neural crest development for fate specification of the iridophore lineage from a multipotent pigment cell progenitor
- Authors
- Petratou, K., Spencer, S.A., Kelsh, R.N., Lister, J.A.
- Source
- Full text @ PLoS One
|
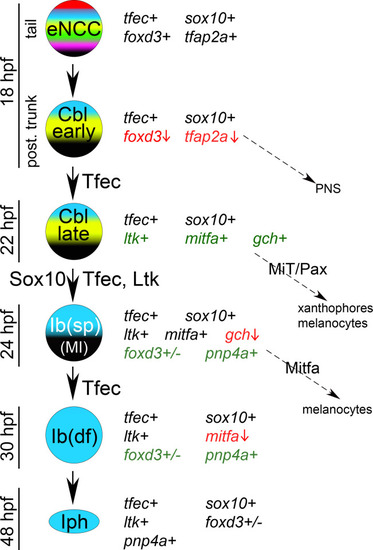
Schematic representation of the previously described model of iridophore generation from the NC, along with the expression characteristics and potential fate choices of individual arising cell types [ |
|
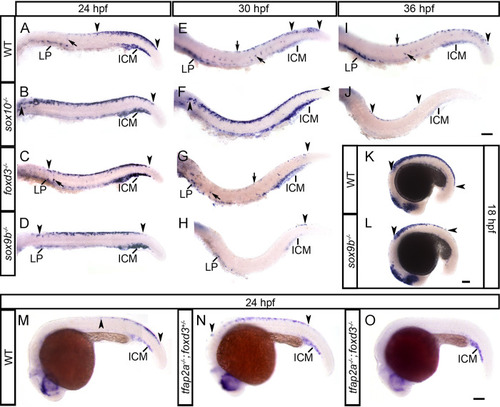
(A–D) Chromogenic whole-mount EXPRESSION / LABELING:
|
|
(A) Schematic showing the distribution of the 8 exons of |
|
(A,B) |
|
(A-F) |
|
(A-J) Peripheral nervous system derivatives develop normally in |
|
In EXPRESSION / LABELING:
|
|
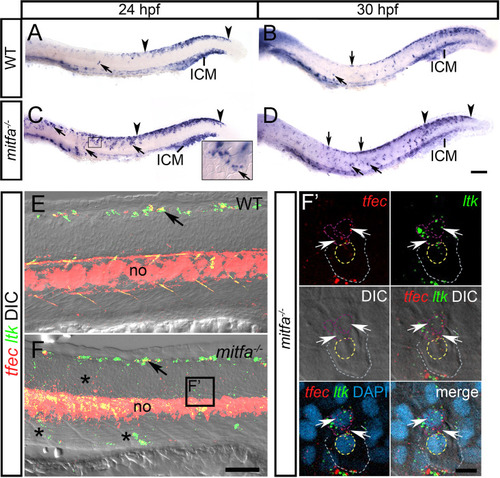
In WT embryos at 24 hpf (A) and at 30 hpf (B), |
|
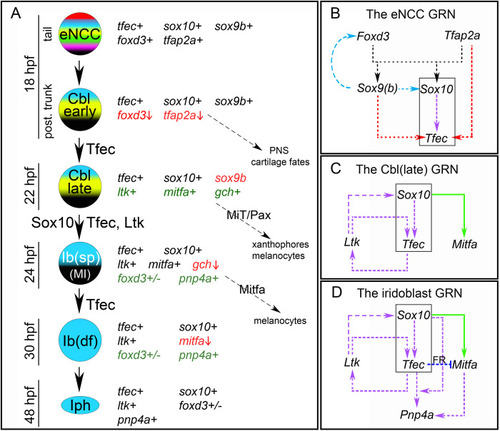
(A) Schematic representation of partially restricted iridophore progenitors during development, along with the expression characteristics and potential fate choices of each ([ |