- Title
-
Atypical Chemokine Receptor 3 Generates Guidance Cues for CXCL12-Mediated Endothelial Cell Migration
- Authors
- Tobia, C., Chiodelli, P., Barbieri, A., Buraschi, S., Ferrari, E., Mitola, S., Borsani, G., Guerra, J., Presta, M.
- Source
- Full text @ Front Immunol
|
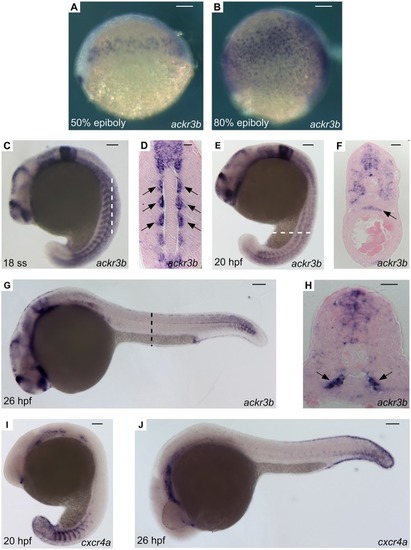
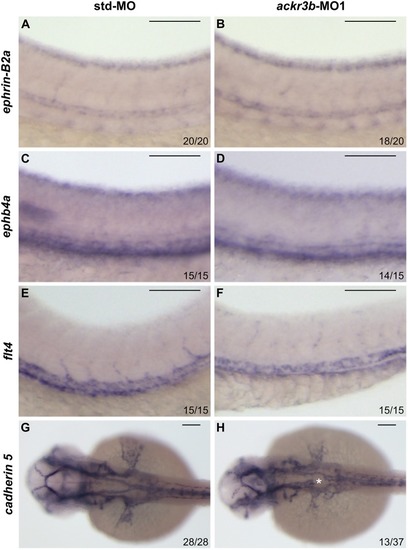
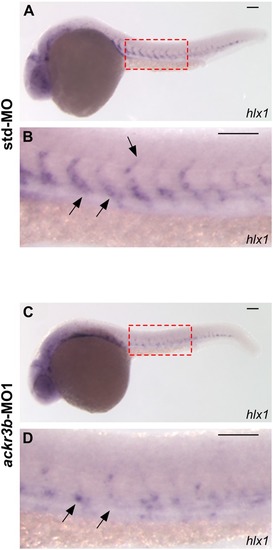
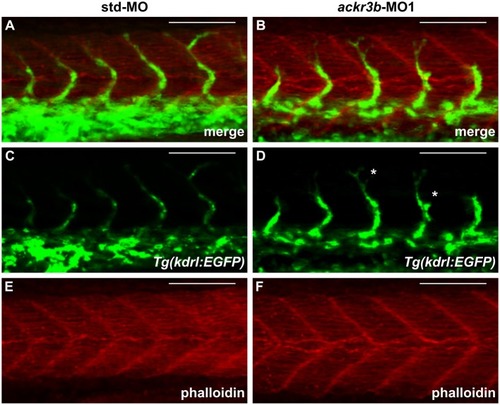
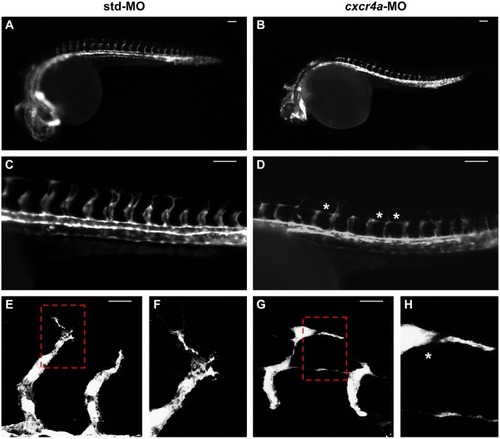
Effect of ackr3b knockdown on zebrafish embryo development. A-E Embryos injected with 0.2 pmoles/embryo of std-MO (A) or ackr3b-MO1 (B-E) were photographed at 28 hpf. Embryos with close-to-normal (B) and mild (C) phenotype were used for all further analyses whereas severe (D) and very severe (E) phenotypes were discarded. F, G Effect of ackr3b knockdown on neuromast migration. WISH analysis of claudin b expression was performed at 48 hpf on zebrafish embryos injected with std-MO (F) or ackr3b-MO1 (G) to assess actual ackr3b knockdown. Note the impaired neuromast migration in ackr3b morphants (arrows in G) when compared to the normal positioning observed in control embryos (arrows in F). The number of embryos presenting the showed phenotype in respect to the total number of analyzed embryos is shown in both panels. |
|
WISH analysis of EXPRESSION / LABELING:
|
|
Effect of |
|
|
|
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
PHENOTYPE:
|
|
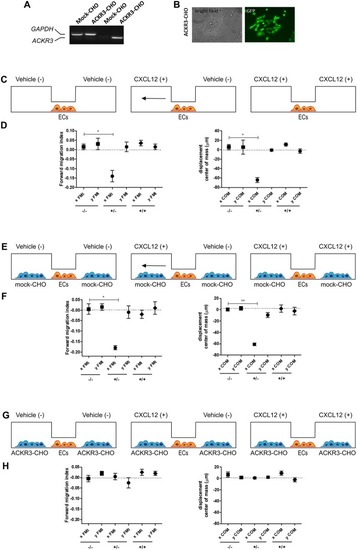
Setup of the μ-slide cell co-culture chemotaxis assay. |

Unillustrated author statements |