- Title
-
A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for Endothelin-1 expression
- Authors
- Nissen, R.M., Amsterdam, A., and Hopkins, N.
- Source
- Full text @ BMC Dev. Biol.
|
The anterior arch mutants. A, C, E, G, I, K), morphology at 4dpf. B, D, F, H, J, L), flat mounted alcian blue stained pharyngeal cartilages. Red asterisks indicate the location of the joint between the M and PQ cartilages. Black asterisks indicate the joint between the CH and hyosymplectic cartilages. A, B) wild type. C, D) HM reduction mutant womhi2914. E, F) HM reduction allele sec61ahi1058. G, H) HM reduction allele sec61ahi2839B. I, J) anterior arch mutant csnk1a1hi2069. M* and CH* are readily identified but dysmorphic. K, L) Meckel's and PQ reductions in mutant dyshi3812. Most homozygous dys/dys animals show 'strong' reductions in the PQ such that only a posterior region of PQ is readily identifiable. M cartilage and the SY region are also strongly reduced. Joint fusion of the CH and HM region are also often present. M, N, O) Typically less than 5% of dys/dys animals show 'mild' reductions or joint fusions. M) Mild dys/dys animal showing only M-PQ and CH-HM joint fusions. N) Mild dys/dys animal showing intermediate PQ splitting in the presence of a well formed M that is fused to the anterior region of the PQ. O) Mild dys/dys animal showing complete PQ splitting but identifiable anterior and posterior PQ regions. The nos/foxI1 and laz/pbx4 mutants have been previously described and are therefore not shown. |
|
The dyshi3812 mutant is a null. A) Diagram indicating the location of the dyshi3812 proviral insert in the first exon of the wdr68 gene. B) RT-PCR analysis of wild type siblings and dys/dys animals showing that wild type wdr68 transcripts are not detectable in the dys mutants indicating that the mutant is a null allele. C) RT-PCR analysis reveals a maternal supply of wdr68 transcripts. cDNA from wild type embryos at zero = unfertilized, 3 hpf, 6 hpf, 10 hpf, and 19 hpf. As negative control, edn1 transcripts are only detected after 10 hpf. Transcripts for the ribosomal gene rpL35 are present at all stages examined. EXPRESSION / LABELING:
|
|
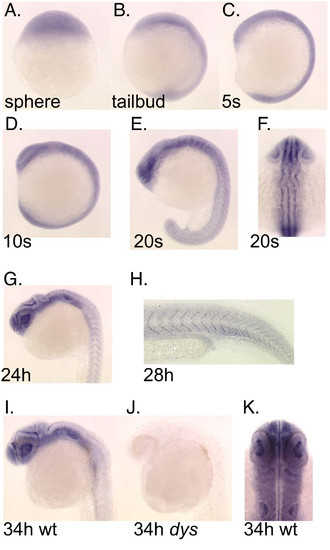
Expression pattern for the wdr68 gene during early development. A) Transcripts are present in all cells of the animal at sphere stage. B) Ubiquitous expression at tailbud stage. C) Ubiquitous expression at 5 somites stage. D) Ubiquitous expression at 10 somites stage. E) Ubiquitous expression with modest enrichment in the forebrain and hindbrain at 20 somites stage. Also note emerging patterned expression in the developing somites. F) Dorsal view of same 20 somites stage animal indicating ubiquitous expression. G) Ubiquitous expression with moderate enrichment in the head region at 24 hpf. H) Higher magnification image of a 28 hpf animal to show patterned expression in somites. I, J) Wild type sibling and dys/dys homozygous mutant at 34 hpf. I) Enriched expression in the head region with much lower expression in somites in a 34 hpf wild type sibling animal. J) Absence of detectable wdr68 transcripts in the dys/dys homozygous mutant sibling. K) Dorsal view of head region of 34 hpf animal showing near ubiquitous expression in developing head structures. |
|
wdr68 activity is required for all 1st arch cartilage formation. A), C), E), G) Lateral views of Alcian blue stained animals at 4dpf. B), D), F), H) Ventral views of Alcian blue stained animals at 4dpf. In panels D, F, and H the posterior arches were removed to enhance clarity for observing the anterior arch defects. A), B) Wild type sibling. C), D) Representative 'strong' phenotype dys/dys animal similar to that shown flatmounted in figure 1 panel D with incomplete loss of M and PQ cartilages. The wdr68MO when injected into wild type animals yields a phenotype very similar to that shown here in panels C and D. E), F) Injecting dys/dys animals with antisense wdr68MO causes complete loss of the PQ, M and CH but does not substantially reduce the HM region. G), H) Representative edn1MO knockdown animal lacking the M and CH but retaining the PQ and HM regions. PHENOTYPE:
|
|
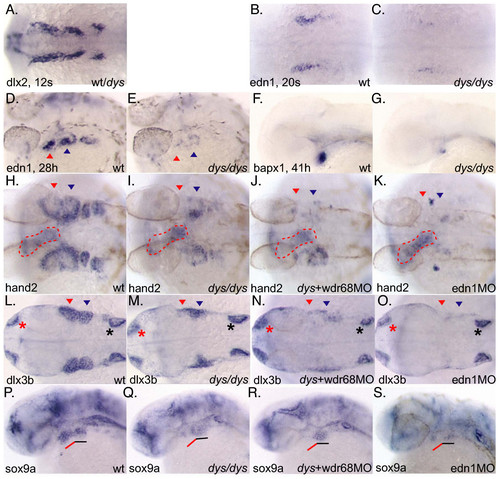
wdr68 acts upstream of the edn1 pathway. A) In situ hybridization (ISH) with dlx2 on 12 somites stage embryos. Wild type and dys/dys mutant animals were indistinguishable. B) ISH on 20 somites stage wild type sibling animal with edn1 probe. C) dys/dys homozygotes display reduced edn1 expression at 20 somites stage. D) ISH on 28 hpf wild type sibling animal with edn1 probe. E) dys/dys homozygotes display reduced edn1 expression at 28 hpf. F) ISH on 41 hpf wild type sibling with bapx1 probe. G) dys/dys homozygotes display reduced bapx1 expression. H, I, J, K, L, M, N, O) Red arrowhead indicates 1st arch expression domain, blue arrowhead indicates 2nd arch expression domain. H) ISH on 28 hpf wild type sibling with hand2 probe. I) dys/dys homozygotes display reduced 1st arch hand2 expression, red arrowhead. Red dotted outline indicates expression in developing heart. J) dys+wdr68MO animals display reduced hand2 expression in both 1st and 2nd arches, red and blue arrowheads, respectively. K) edn1-MO animals lack hand2 expression in both the 1st and 2nd arches. L, M, N, O) Red asterisk indicates olfactory expression domain. Black asterisk indicates ear expression domain. L) ISH on 28 hpf wild type sibling with dlx3b probe. M) dys/dys homozygotes display reduced 1st arch dlx3b expression, red arrowhead. N) dys+wdr68MO animals display reduced dlx3b expression in both 1st and 2nd arches, red and blue arrowheads, respectively. O) edn1-MO animals lack dlx3b expression in both the 1st and 2nd arches. P, Q, R, S) Red underline indicates 1st arch region for sox9a expression. Black underline indicates 2nd arch region for sox9a expression. P) ISH with sox9a probe on 28 hpf wild type sibling. Q) Reduction of 1st arch sox9a expression without significant effect on 2nd arch sox9a expression in dys/dys animals. R) Injecting dys animals with wdr68MO caused further reduction of 1st arch sox9a expression without substantial effects on 2nd arch sox9a expression. S) edn1-MO animals show reduced but still detectable sox9a expression in both the 1st and 2nd arches. EXPRESSION / LABELING:
|
|
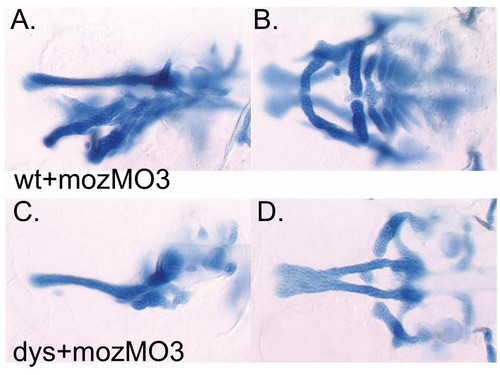
Transformation of 2nd arch identity transforms 2nd arch wdr68 sensitivity. A, C) Lateral views at 4dpf. B, D) Ventral views at 4dpf. A, B) Wild type animal injected with mozMO transforms 2nd arch cartilage elements into mirror image duplicates of 1st arch cartilages. C, D) Injecting dys animals with mozMO reveals similar cartilage losses as in maternal wdr68 knockdowns shown in (Fig. 4E, F) but with further reduction of HM region presumably through mirror image duplication of the missing 1st arch cartilages. PHENOTYPE:
|
|
The Wdr68 protein co-localizes with Dyrk1a. Transiently transfected HEK-293FT cells. A, E, I) Fluorescence of GFP and fusion proteins. B, F, J) Fluorescence of mRFP1 and fusion proteins. C, G, K) Fluorescence of nuclei stained with DAPI. D, H, L) Composite overlay image of GFP, RFP and DAPI signals. A, B, C, D) GFP and mRFP1 distribute throughout the cytoplasm and nucleus. E, F, G, H) The GFP-Wdr68 fusion and the mRFP1-Dyrk1a fusion are co-localized to the nucleus. I, J, K, L) The GFP-T284F Wdr68 mutant does not co-localize with nuclear mRFP1-Dyrk1a. |
|
wdr68 activity is functionally conserved. A) Sequence similarity scores generated using ClustalW between wdr68 homologs among selected representative organisms. Region shown contains the functionally important position, highlighted in red, identified in the ttg1-9 allele. B) The T284F mutant cannot rescue the dys mutant phenotype. Live 4dpf animal shown. C) The drosophila homolog CG14614 rescues the dys mutant phenotype as efficiently as the zebrafish transcripts. D) Alcian blue staining of attempted rescue animals using T284F transcripts. E) Alcian blue staining of animals rescued using the drosophila homolog CG14614. F) Chart summarizing the results of a rescue experiment showing that zebrafish wdr68 transcripts rescue the strong mutant phenotype, that T284F transcripts fail to rescue, and that CG14614 rescues as efficiently as zebrafish wdr68. |
|
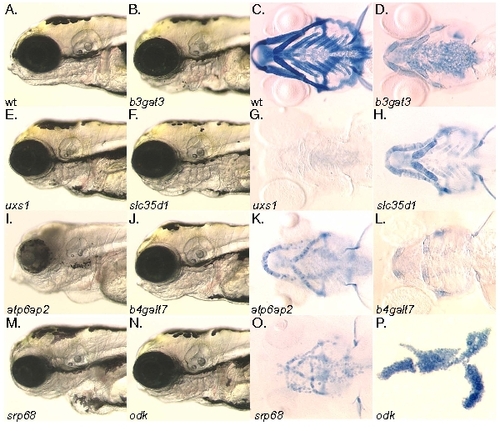
The chondrocyte differentiation and cell morphology class mutants. A, B, E, F, I, J, M, N) Head morphology at 4dpf. C, D, G, H, K, L, O, P) Alcian blue stained pharyngeal cartilages at 4dpf viewed ventrally, except for panel (P) which is flat mounted for comparison with figure 1. A, C) Wildtype. B, D) b3gat3hi307 mutant. E, G) uxs1hi954 mutant. F, H) slc35d1hi3378 mutant. I, K) atp6ap2hi3681 mutant. J, L) b4galt7hi4063A Ehlers-Danlos mutant. M, O) srp68hi4153 mutant. N, P) odkhi1042 cell morphology mutant. The jef/sox9a, goz/mbtps1, kny/glp6 mutants have been previously described and so are omitted here. |
|
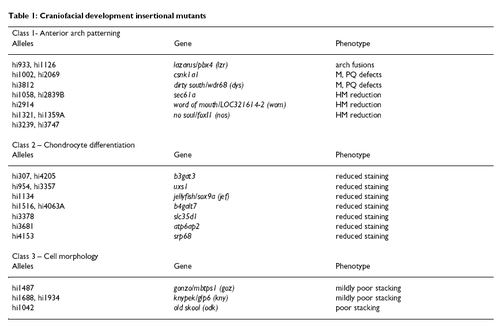
Craniofacial development insertional mutants PHENOTYPE:
|

Unillustrated author statements |