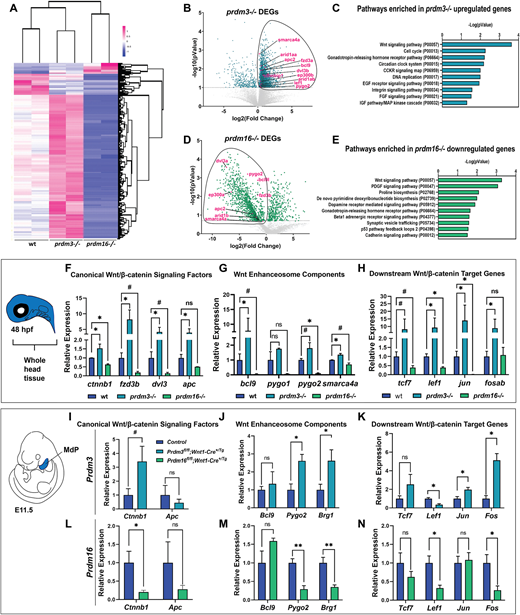
Fig. 3
Fig. 3 Prdm3 and Prdm16 differentially regulate Wnt/β-catenin signaling components in cranial NCCs. (A-E) RNA-seq was performed on sox10:EGFP FACS-sorted cranial NCCs at 48 hpf from wild-type, prdm3−/− and prdm16−/− zebrafish. (A) Heatmap showing z-scaled FPKM values for significant genes from prdm3−/− and prdm16−/− mutants. (B,D) Volcano plots showing the distribution of DEGs in prdm3−/− (B) and prdm16−/− (D), including Wnt/β-catenin signaling components (highlighted in magenta). (C,E) The upregulated genes in prdm3 mutants and downregulated genes in prdm16 mutants (circled in B and D) were subjected to GO (PANTHER) Pathway Enrichment Analysis. (F-H) Validation of RNA-seq transcriptomic changes on RNA isolated from whole heads pooled from five to seven embryos per genotype; mean±s.d. RT-qPCR was performed for selected Wnt/β-catenin signaling component genes, including canonical Wnt/β-catenin signaling factors (ctnnb1, fzd3b, dvl3, apc) (F); Wnt enhanceosome transcriptional complex members (bcl9, pygo1, pygo2, smarca4a) (G); and downstream Wnt/β-catenin target genes (tcf7, lef1, jun, fosab) (H). (I-N) Validation of zebrafish RNA-seq transcriptomic changes in E11.5 mouse mandibular processes. RT-qPCR was performed for selected genes, including canonical Wnt/β-catenin signaling factors (Ctnnb1, Apc) in Prdm3 (I) and Prdm16 (L) mutants; Wnt enhanceosome transcriptional complex members [Bcl9, Pygo2, Brg1 (Smarca4a)] in Prdm3 (J) and Prdm16 (M) animals; and downstream Wnt/β-catenin target genes (Tcf7, Lef1, Jun, Fos) in Prdm3 (K) and Prdm16 (N) mutants. n=3 per genotype; mean±s.d. MdP, mandibular process. *P≤0.05, **P≤0.005, #P≤0.1; ns, not significant (unpaired, two-tailed Student's t-test).