Fig. 5
- ID
- ZDB-FIG-250722-6
- Publication
- Evans et al., 2025 - Transcriptome remodelling and changes in growth and cardiometabolic phenotype result following Grb10a knockdown in the early life of the zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
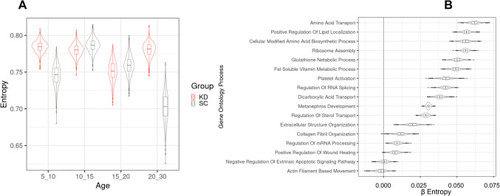
Hypergraph entropy in the transcriptome reveals differences over time and between functions. ( |