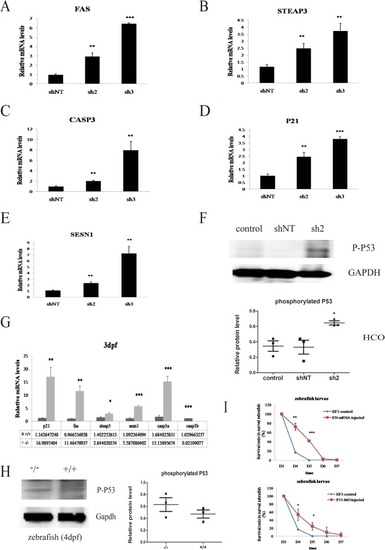
The p53 pathway mediates cell development in HCO cells and zebrafish. a Expression of the FAS gene in cells transfected with different lentiviruses (sh2, sh3, and shNT as a scramble control) at 3 days before cell confluence. HCO cells subjected to EFTUD2 knockdown (sh2 and sh3) exhibited increased FAS levels. b The expression of CASP3 in the sh2 and sh3 groups was dramatically increased compared with that in the shNT group. cSTEAP3 mRNA levels in the sh2 and sh3 groups were markedly higher than that in the negative control. dP21 mRNA levels were also highly increased in the sh2 and sh3 groups. eSESN1 was highly expressed in the sh2 and sh3 groups. f HCO cells transfected with sh2 had a higher expression of phosphorylated P53 protein (53 kDa) than the nontransfected (control) and shNT groups. g Expression of relevant genes involved in the P53 pathway in the EFTUD2 knockout (−/−) and WT (+/+) zebrafish. h The expression of phosphorylated P53 in EFTUD2-mutated larvae (−/−) with curved bodies and WT larvae at 4 dpf. i The survival rate among the curved F3 generation hybridizing from EFTUD2 heterozygous mutants (EF3 control), EF3 controls injected with EFTUD2 normal mRNA (EN mRNA), and p53 morpholino (P53-MO) during the early developmental stage (*P < 0.05, **P < 0.01, and ***P < 0.001)
|

