- Title
-
The Antioxidant Drug Edaravone Binds to the Aryl Hydrocarbon Receptor (AHR) and Promotes the Downstream Signaling Pathway Activation
- Authors
- Veroni, C., Olla, S., Brignone, M.S., Siguri, C., Formato, A., Marra, M., Manzoli, R., Macario, M.C., Ambrosini, E., Moro, E., Agresti, C.
- Source
- Full text @ Biomolecules
|
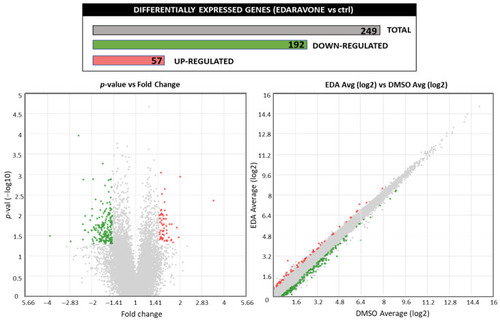
Effect of EDA treatment on the OPC transcriptome. Purified OPCs were incubated with 100 µM EDA or vehicle alone (DMSO) for 14 h. RNA was extracted, reverse transcribed and subjected to targeted transcriptome analysis. Treatment with EDA regulated the expression of 249 genes ranging in a ±1.5-fold change with a |
|
Validation of the effect of EDA treatment on AHR-related transcript expression in OPCs. OPCs were treated with EDA 30 µM, 100 µM or vehicle (DMSO) alone for 14 h. Total RNA was extracted and reverse transcribed and then the expression of the selected genes was evaluated using qPCR. Data are expressed as 2−ΔΔCt relative to the housekeeping gene |
|
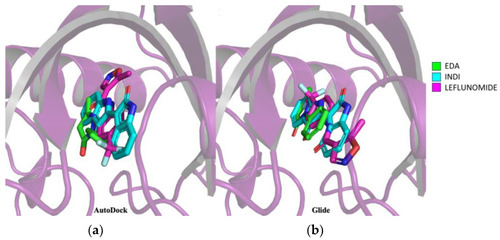
Prediction of EDA–AHR binding mode by molecular docking. ( |
|
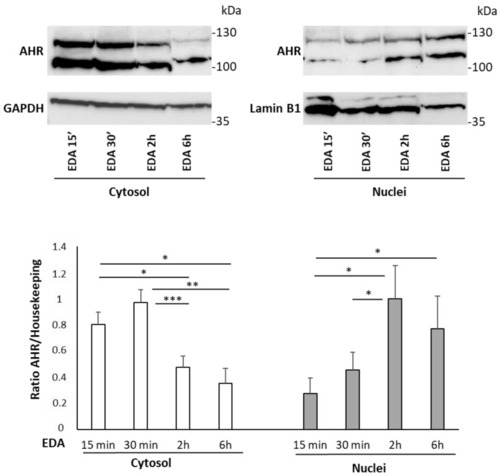
EDA induction of AHR nuclear translocation in the SH-SY5Y neuroblastoma cell line. SH-SY5Y human neuroblastoma cells were incubated with 100 µM EDA for 15 min, 30 min, 2 h and 6 h. The cytosolic and nuclear fractions were separated and the expression level of AHR in each fraction was evaluated by Western blot analysis. GAPDH and LAMINB1 were used for protein content normalization in cytosol and nuclei, respectively. Bars represent the mean ± SEM of 4 experiments. * |
|
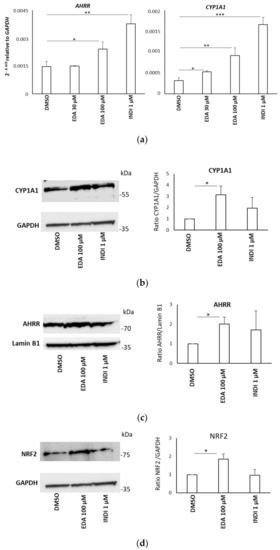
EDA induction of AHR target genes in the SH-SY5Y neuroblastoma cell line. ( |
|
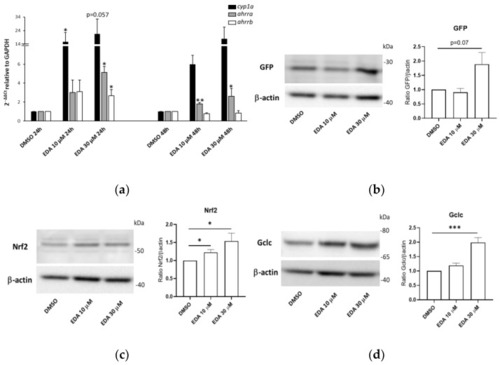
EDA promotes AHR and NRF2 pathway activation in zebrafish larvae. ( |
|
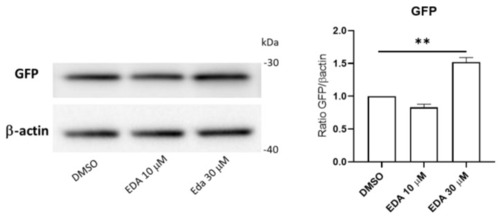
EDA treatment induces reporter expression in Olig2 transgenic fish. Representative Western blot for the eGFP reporter protein on fish trunk whole lysates from control DMSO and EDA-treated |
|
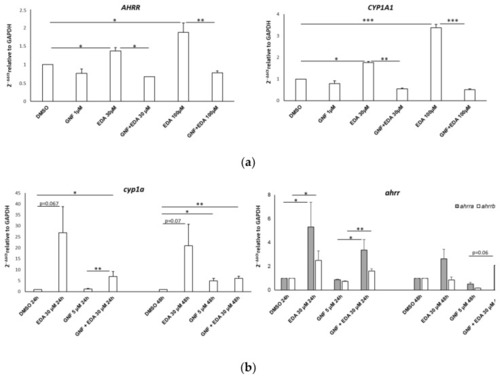
AHR inhibition curtails EDA-mediated AHR target gene upregulation in vitro and in vivo. Bar graphs show the gene expression levels detected by qPCR on RNA obtained from SH-SY5Y cells ( |