- Title
-
Transcriptome analysis of Edwardsiella piscicida during intracellular infection reveals excludons are involved with the activation of a mitochondrion-like energy generation program
- Authors
- Lanza, A., Kimura, S., Hirono, I., Yoshitake, K., Kinoshita, S., Asakawa, S.
- Source
- Full text @ MBio
|
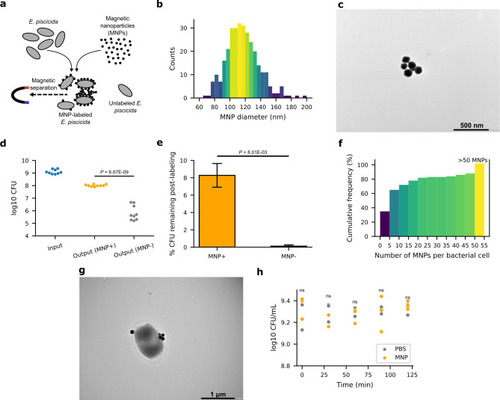
MNPs bind to the surface of |
|
MNP-labeled |
|
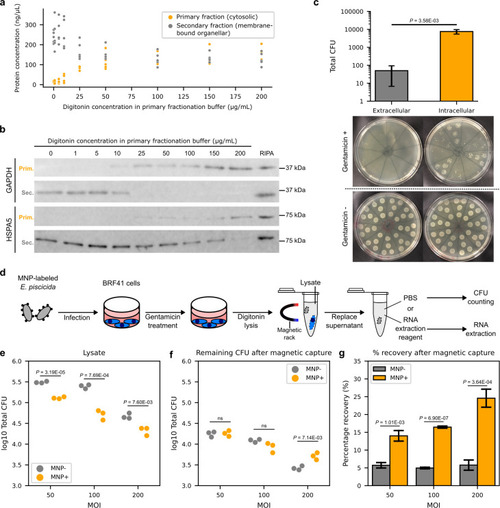
Enrichment of |
|
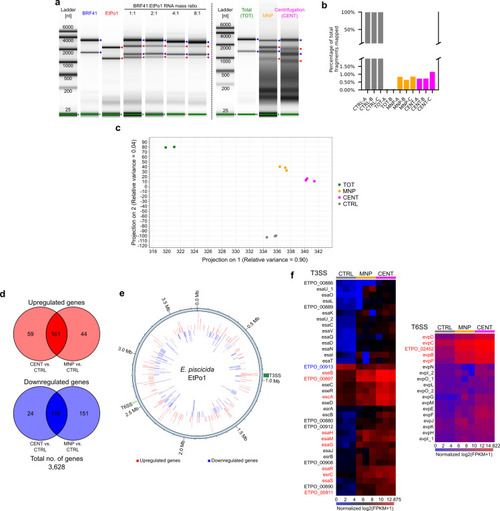
Bacterial sugar transport and glycolysis do not constitute the preferred carbon and energy source for |
|
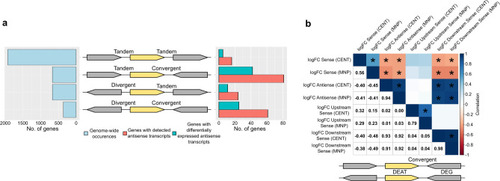
Orientational contexts of genes on the |
|
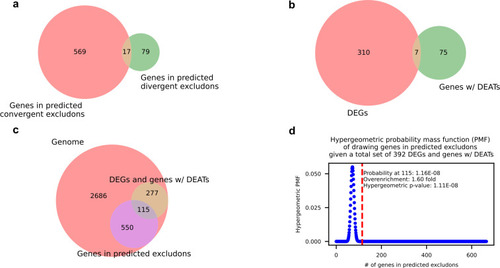
Genes in predicted excludons are enriched in the set of genes with perturbed expression during intracellular infection. ( |
|
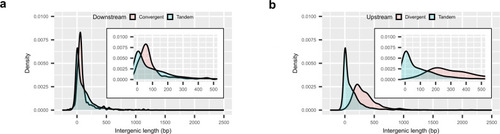
Intergenic distances between convergent and divergent gene pairs in the |
|
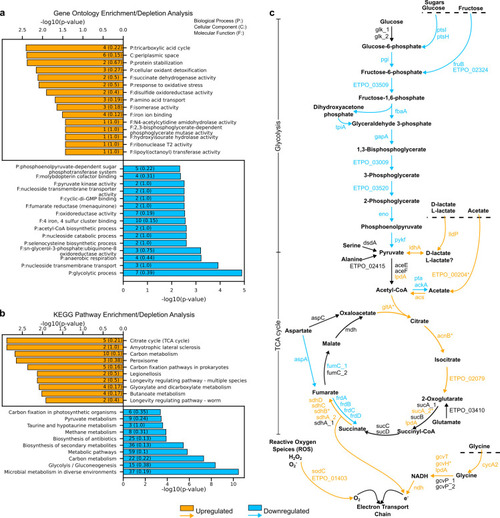
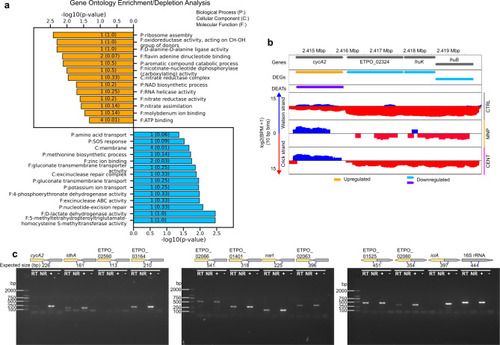
Potential roles and RT-PCR validation of predicted excludons. (a) GO enrichment analysis of responder genes in convergent excludons. Orange and blue bars represent enriched annotations for upregulated and downregulated antisense transcripts, respectively. Numbers on the bars represent the number of genes with the given annotation, and numbers in parentheses represent the ratio of genes to all genes with the annotation. (b) Representative responder gene in convergent excludon, |