- Title
-
Staphylococcus aureus cell wall structure and dynamics during host-pathogen interaction
- Authors
- Sutton, J.A.F., Carnell, O.T., Lafage, L., Gray, J., Biboy, J., Gibson, J.F., Pollitt, E.J.G., Tazoll, S.C., Turnbull, W., Hajdamowicz, N.H., Salamaga, B., Pidwill, G.R., Condliffe, A.M., Renshaw, S.A., Vollmer, W., Foster, S.J.
- Source
- Full text @ PLoS Pathog.
|
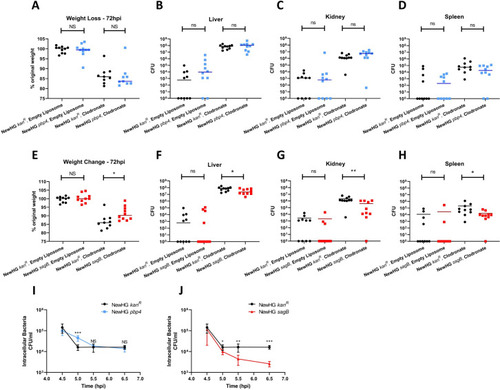
Mice (n = 10) were injected with approximately 1 x 105 CFU of NewHG |
|
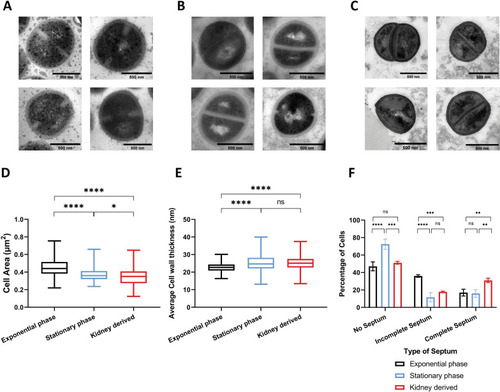
Thin sections of chemically fixed |
|
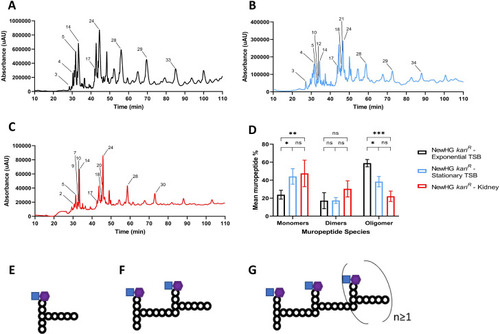
Representative muropeptide profiles of NewHG |
|
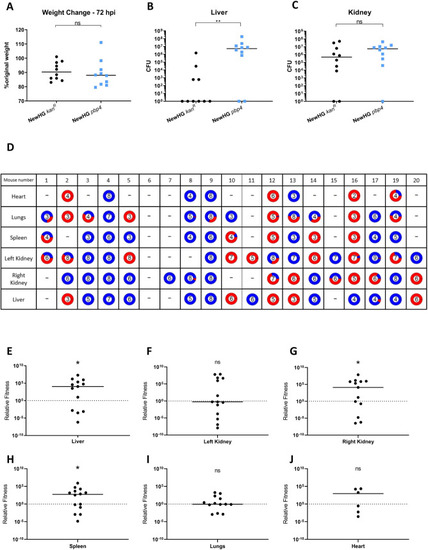
Mice (n = 10) were injected with approximately 5x106 CFU |
|
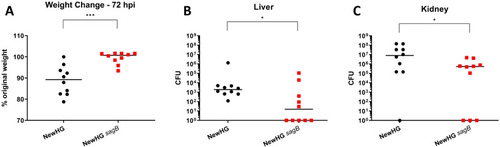
Mice (n = 10) were injected with approximately 5x106 CFU |
|
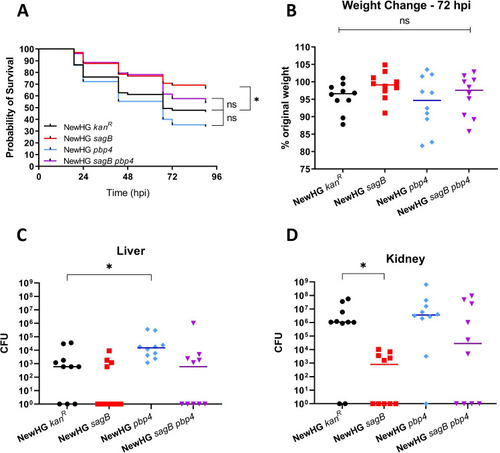
Approximately 1 x 107 CFU of |
|
PHENOTYPE:
|
|
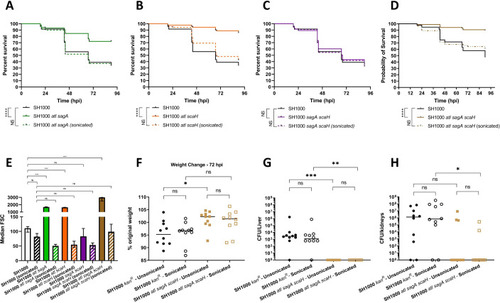
Survival curves comparing the virulence of 1300 CFU parental NewHG (SJF 3663, black lines) to |