- Title
-
MITF reprograms the extracellular matrix and focal adhesion in melanoma
- Authors
- Dilshat, R., Fock, V., Kenny, C., Gerritsen, I., Lasseur, R.M.J., Travnickova, J., Eichhoff, O.M., Cerny, P., Möller, K., Sigurbjörnsdóttir, S., Kirty, K., Einarsdottir, B.Ó., Cheng, P.F., Levesque, M., Cornell, R.A., Patton, E.E., Larue, L., de Tayrac, M., Magnúsdóttir, E., Helga Ögmundsdóttir, M., Steingrimsson, E.
- Source
- Full text @ Elife
|
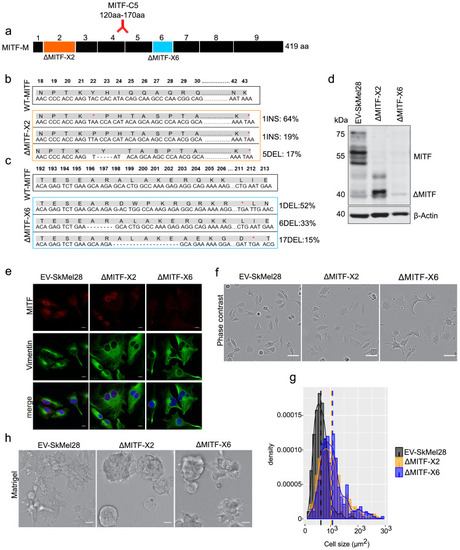
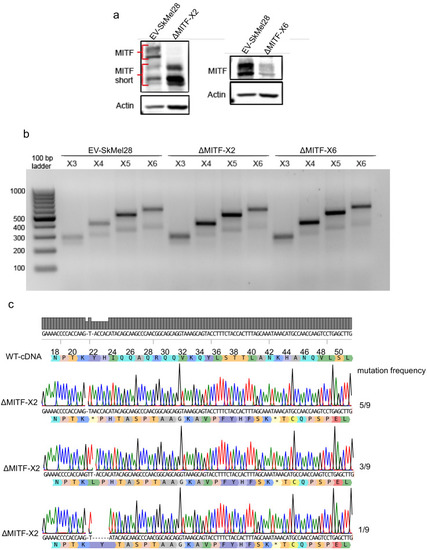
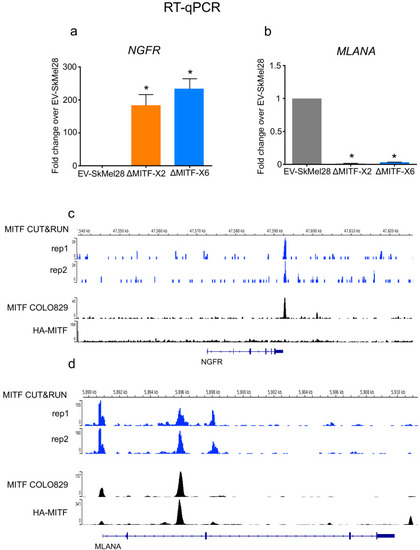
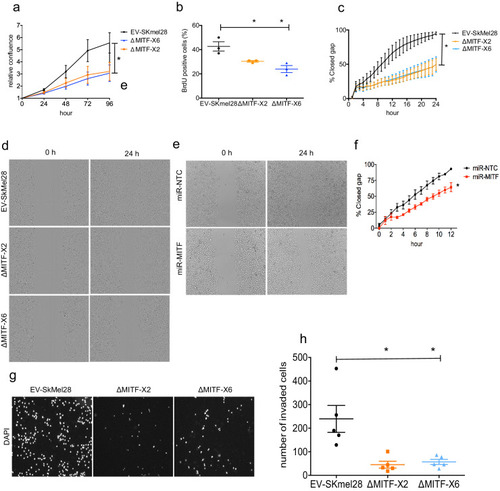
MITF depletion affects cell size and cell-matrix interaction. (a) Schematic illustration of MITF-M isoform and gRNA targeted location at exon 2 and exon 6. The epitope location for MITF C5 antibody spanning exons 4 and 5 is shown. (b, c) Mutations detected in ?MITF-X2 and ?MITF-X6 cell lines; amino acid sequence numbering was indexed relative to MITF-M. Percentage of mutations was derived from WGS analysis by counting sequenced fragments aligned to the mutated regions. (d) Western blot showing the MITF band in EV-SkMel28, ?MITF-X2, and ?MITF-X6 cell lines. (e) Immunostaining for MITF and Vimentin in EV-SkMel28, ?MITF-X2, and ?MITF-X6 cell lines, scale bar (10 µm). (f, g) Phase-contrast microscopy and cell size quantification using Image J with at least 200 images taken for both MITF-KO and EV-SkMel28 cell lines, scale bar (10 µm). Average cell size for each cell line is indicated by dashed lines; EV-SkMel28 cells (6502 µm2, SEM: 460), ?MITF-X2 (10,395 µm2, SEM: 270) and the ?MITF-X6 (10,825 µm2, SEM: 330). (h) Bright-field images of MITF-KO and EV-SkMel28 cells grown on top of matrigel, scale bar (10 µm). |
|
( |
|
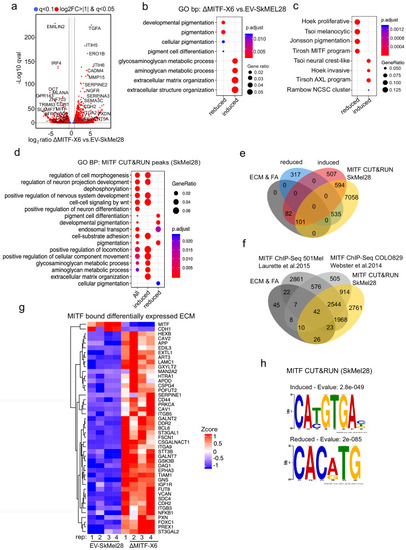
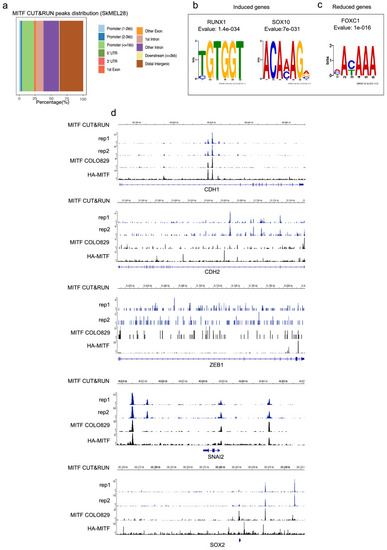
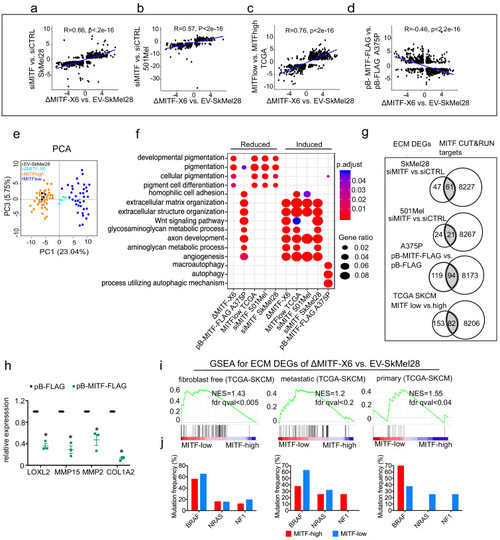
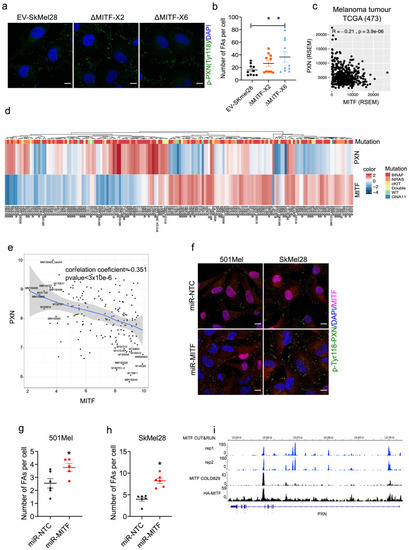
MITF binds and represses genes of extracellular matrix (ECM) and focal adhesion genes. (a) Volcano plot showing 2136 DEGs with qval <0.5 among which 1516 genes with log2FC?|1| fold change in expression ?MITF-X6 vs. EV-SkMel28. (b) GO BP analysis of the 1284 induced and 852 reduced DEGs between ?MITF-X6 vs. EV-SkMel28 cells presented in dot plot; adjusted p-value is red lowest to blue highest; gene ratio is the ratio between DEGs and all genes in the GO category. (c) Dot plot of enrichment analysis showing the enrichment of gene signatures from the literature in reduced and induced DEGs of ?MITF-X6 vs. EV-SkMel28. p Value is red lowest to blue highest; gene ratio is the ratio between genes and all genes in the GO category; reduced: genes reduced in expression in ?MITF-X6 compared to EV-SkMel28; induced: genes induced in expression in the ?MITF-X6 compared to EV-SkMel28. (d) GO BP analysis of MITF CUT and RUN peaks associated genes were plotted using Clusterprofiler (Yu et al., 2012) in R; All: MITF CUT and RUN peak-associated genes, induced and reduced: Induced or reduced DEGs of ?MITF-X6 vs. EV-SkMel28 cells based on MITF CUT and RUN peak presence on their gene promoter or distal region binding. (e) Venn diagram showing the overlap between MITF targets identified from MITF CUT and RUN with induced, reduced, ECM and focal adhesion DEGs of ?MITF-X6 vs. EV-SkMel28 cells. (f) Venn diagram displaying the common overlap between MITF ChIP-seq targets in different cell lines and differentially expressed ECM and focal adhesion genes in ?MITF-X6 vs. EV-SkMel28 cells. (g) Heatmap showing the differentially expressed ECM genes in ?MITF-X6 vs. EV-SkMel28 cells that are commonly bound by MITF across different MITF CUT and RUN data sets. Zcore converted TPM value from RNA-seq data was used for plotting. (h) Motif analysis of MITF CUT-and-RUN targets of induced and reduced genes in ?MITF-X6 vs. EV-SkMel28 cells. |
|
( |
|
( |
|
( |
|
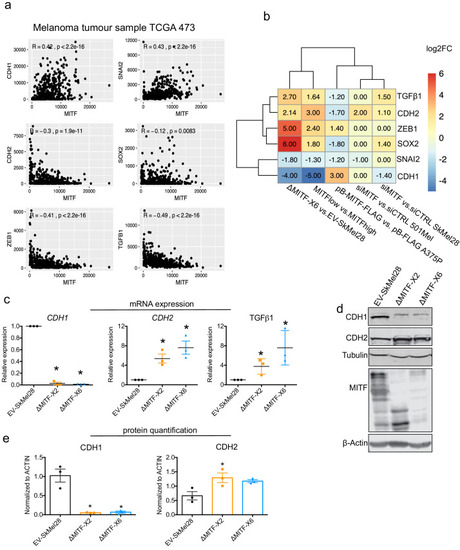
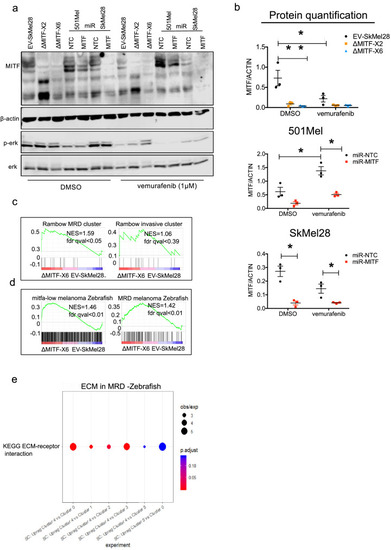
Epithelial-to-mesenchymal transition (EMT) genes are directly regulated by MITF. (a) Scatter plot displaying the Spearman correlation between MITF mRNA expression with EMT genes in the 472 melanoma tumor samples from TCGA; MITF displayed positive correlation with CDH1 and SNAI2 and negative correlation with ZEB1, TGFß1, and CDH2. (b) Differentially expressed EMT genes plotted as heatmap using the log2 fold change value of DEGs of MITF depletion in SkMel28 and 501Mel cells, MITF overexpression in A375P cells and MITFlow&high melanoma tumors. (c) Real-time qPCR (RT-qPCR) evaluation of EMT genes in the EV-SkMel28, ?MITF-X2, and ?MITF-X6 cell lines. Fold change in the expression calculated over EV-SkMel28. Error bar represents standard error of the mean (* p value <0.05) was calculated using one-way ANOVA (multiple correction with Dunnett test). (d, e) Western blot analysis and quantification (Fiji Image J) of protein expression of CDH1, CDH2, and MITF in EV-SkMel28, ?MITF-X2, and ?MITF-X6 cell lines. ß-Actin was used as loading control. * p value <0.05 was calculated by one-way ANOVA (multiple correction with Dunnett test). |
|
(
|
|
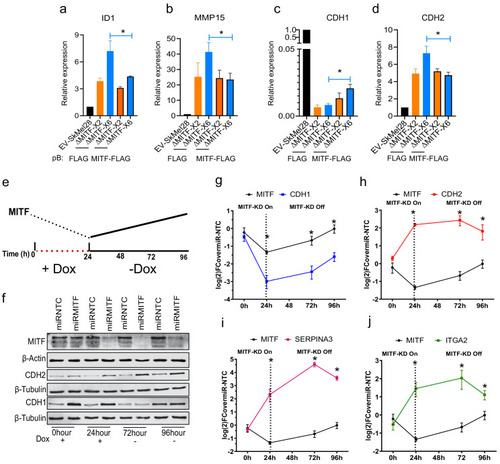
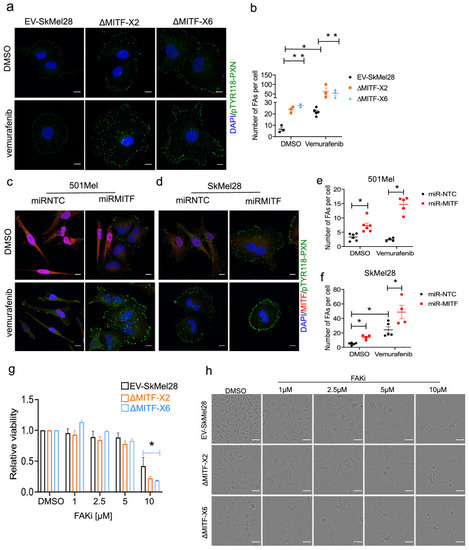
MITF mediates formation of focal adhesion. (a?b) Immunostaining for p-PXNTYR118 and quantification of p-PXNTYR118-positive focal points in EV-SkMel28, ?MITF-X2 and ?MITF-X6 cell lines treated with DMSO (a, upper panel) or vemurafenib (a, lower panel). (c?d) Immunostaining for p-PXNTYR118 and MITF and (e, f) quantification of p-PXNTYR118-positive focal points in miR-NTC and miR-MITF treated 501Mel and SkMel28 cells. Error bars represent standard error of the mean, * p value <0.05 was calculated by two-way ANOVA (multiple correction with Sidak test). (g) Bar plot represents the relative viable cells compared to DMSO control with increasing concentration of FAK inhibitor (PF562271) for 72 hr. Error bars represent standard error of the mean, * p value <0.05 was calculated by two-way ANOVA (multiple correction with Tukey test). (h) Incucyte images of cells after 72 hr treatment with increasing concentrations of FAKi (PF562271) and with respective DMSO control. |
|
(
|
|
( |
|
( |