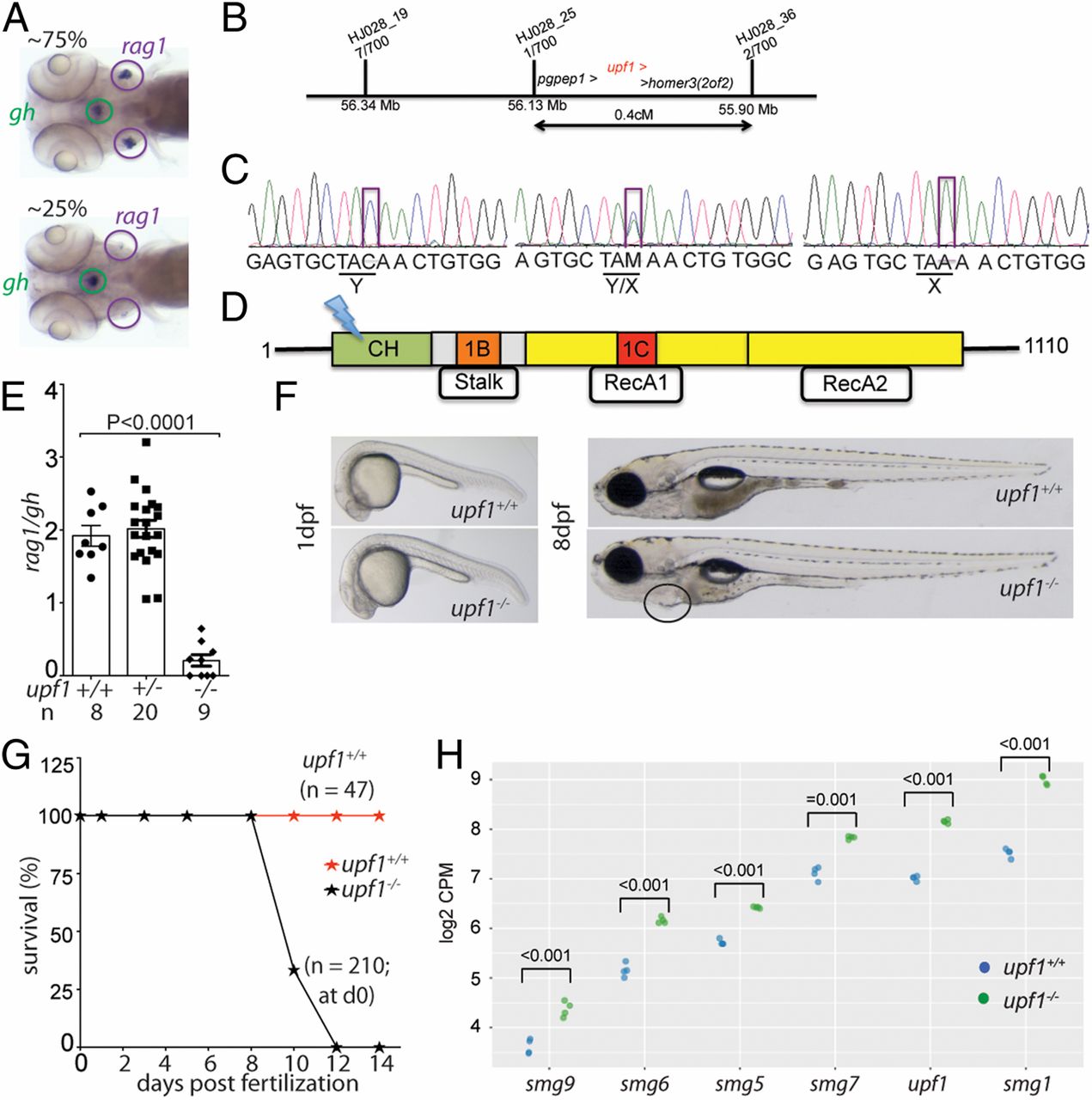
Fig. 1
Fig. 1 Characterization of a upf1 mutant allele. (A) In line HJ028, crosses of mutant carriers generate about 25% of fish with small numbers of rag1-expressing cells in the thymic rudiment, whereas the other siblings appear phenotypically normal. The expression of gh in cells of the hypophysis serves as an internal control. Note that some upf1 mutant fish lack detectable expression of rag1 (compare with quantitative analysis in E). (B) Schematic of the critical region for the HJ028 mutation on chromosome 2, as determined by linkage mapping with polymorphic markers and genome sequencing; the three genes located in the interval are indicated; the numbers of recombinants for each of the polymorphic markers observed in 700 meioses are indicated. (C) Sanger sequencing traces of the three genotypes in the HJ028 line. The C > A transversion is indicated (position 2:56,181,617; Zv11). (D) Schematic of the upf1 protein domains; cysteine- and histidine-rich zinc-finger domain (CH), amino acids 121 to 272; 1B/Stalk: amino acids 325 to 426; 1C: amino acids 567 to 620; RecA1: amino acids 449 to 700; RecA2: amino acids 701 to 918. The stop codon is introduced at amino acid position 163 (Y163X) in the cysteine- and histidine-rich zinc-finger domain. (E) Thymopoietic capacity of upf1 mutants as measured by RNA in situ hybridization using probes specific for rag1 (expressed in developing thymocytes) and gh (expressed in a subset of cells in the hypophysis). The latter probe serves as an internal control in two ways: Technical, to monitor the hybridization process as such; and biological, to ascertain the tissue specificity of the observed genetic effects. Each data point represents one fish; mean ± SEM. (F) Macroscopic appearance of upf1 mtants and their wild-type siblings at two time points. The cardiac region of the mutant is highlighted; the edema manifests itself as a ventral bulge, which is absent in wild-type siblings. (Magnification: 10×.) (G) Impaired survival of upf1 mutants. The survival rate of upf1 mutants was determined by analyzing aliquots of a clutch of initial 210 fish generated by a cross of heterozygous parents; the survival rate of upf1 wild-type fish was determined from a cross of wild-type fish of the same background. (H) Up-regulation of genes encoding components of the NMD pathway in upf1 mutants. Gene-level expression is shown as log2 counts per million (CPM); the levels of significance of differences between the two datasets for each gene are indicated.