- Title
-
An antagonistic role of clock genes and lima1 in kidney regeneration
- Authors
- He, X., Wang, Z., Cheng, L., Wang, H., Sun, Y.
- Source
- Full text @ Commun Biol
|
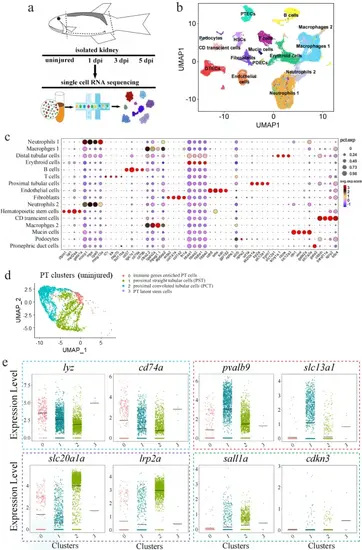
scRNA-seq data analysis of zebrafish kidneys.a Schematics of the study design. b UMAP embedding of scRNA-seq data. Using kidney marker genes, cells were annotated into the indicated 16 major clusters. PTECs: proximal tubular epithelial cells; DTECs: distal tubular epithelial cells; B cells: B lymphocytes; T cells: T lymphocytes; PDECs: pronephric duct cells; PD; podocytes; HSCs: hematopoietic stem cells. c Bubble plot displaying the cluster-enriched marker genes. d UMAP projection of uninjured/wild-type proximal tubular cells (PT), demonstrating four sub-clusters. Subcluster 0: the immune gene-enriched cells; subcluster 1: proximal straight tubular cells (PST); subcluster 2: proximal convoluted tubular cells (PCT); sub-cluster 3: the resident stem cell population. e Violin plots grouped by meta-clusters, demonstrating the subcluster-specific expression of the indicated genes. The dashed colored boxes were used to indicate the markers of the four subclusters. 0–3 of the X-axis: the subclusters 0, 1, 2, 3. |
|
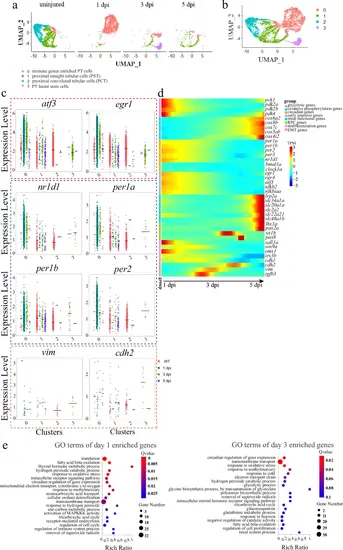
Dynamic gene expression of the 4 PT subclusters during renal regeneration.a UMAP embedding of scRNA-seq data colored by time points highlights the progressive nature of 4 subclusters during regeneration. b UMAP embedding of all cells profiled during early stages of renal regeneration. c Violin plots showing dynamic expression of the indicated genes, grouped into markers of early response, RPCs, circadian clock and EMT. The colored boxes were used to highlight the indicated groups. 0–3 of the X-axis: the subclusters 0–3. d Heat map showing the dynamic expression of the indicated genes, grouped into 8 categories that are shown on the upper right corner. e GO terms of the enriched genes at 1 and 3 dpi. |
|
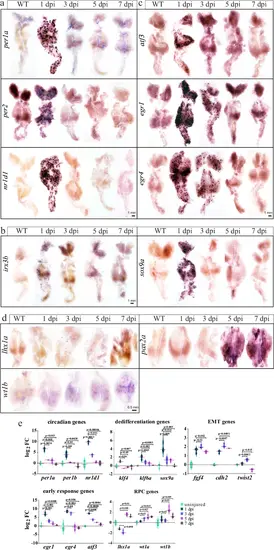
Validation of the dynamic expression of the indicated genes.a WISH image of the indicated circadian clock genes. b WISH images of the indicated dedifferentiation genes. c WISH images of the indicated early response genes. d WISH images of the RPC genes. e Plots showing the dynamic expression of the indicated genes during a time course of 7 days after AKI, based on 3 bulk RNA-seq replicates. Log2 FC: the log2 fold change of the indicated gene’s expression at the indicated time points relative to day 0 (uninjured kidneys). P-values were indicated to show statistical significance. WISH experiments were repeated three times, and shown were representative data. |
|
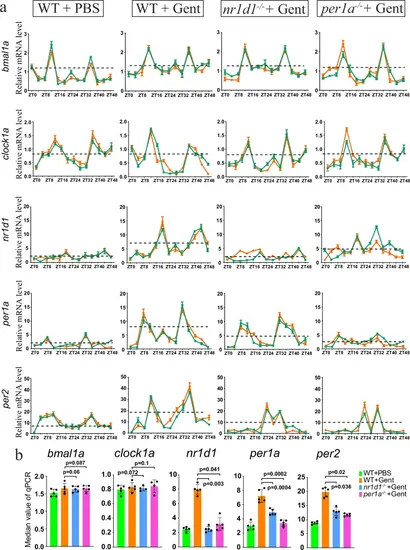
Rhythmic expression of the circadian clock genes.a qRT-PCR results showing the rhythmic expression of the indicated clock genes at 8 h time points over 48 h, with and without gentamicin (Gent) treatment. The green and red colors were used to represent the average values of two replicates (each with three biological repeats). The dashed black lines indicated the median values of mRNA levels. qRT-PCR was repeated at least four times, and shown were two replicates. b Quantification of panel a, based on the median values of mRNA levels. P-values for each panel were indicated to show the statistical significance. |
|
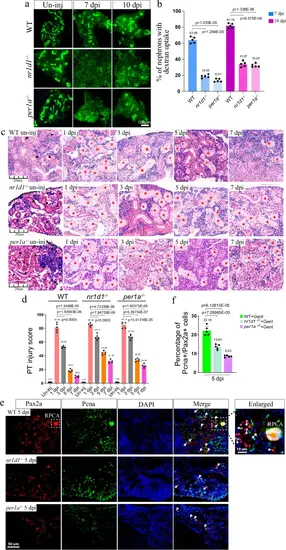
Loss of per1a and nr1d1 leads to renal regeneration defects.a Representative dextran fluorescent signals of kidneys at the indicated time points, in control, per1a−/−, and nr1d1−/− zebrafish. The ability to uptake dextran was used to indicate the regenerated nephrons. b Quantification of panel a (n = 3–5 different regions of each group), showing the percentage of fluorescent nephrons. c HE staining results of kidneys at the indicated time points, in control, per1a−/−, and nr1d1−/− animals. Black arrow heads: normal tubules, and red stars: damaged tubules. d Quantification of panel c (n = 4–5 different regions of each group), to show the renal recovery dynamics after AKI. The PT injury score is used to show the percentage of abnormal PTs. e Pax2a and Pcna double-staining images of kidneys from control, per1a−/−, and nr1d1−/− animals at 5 dpi. White arrows: Pax2a and Pcna double positive single cells. White box with the dashed line: Pax2a expressing RPC aggregates, which were zoomed in on the right corner. f Quantification of panel e (n = 4–5 different regions of each group), with p-values. Gent: gentamicin. The IF and HE were repeated three times, and shown were representative data. p-values for each panel were indicated to show the statistical significance. |
|
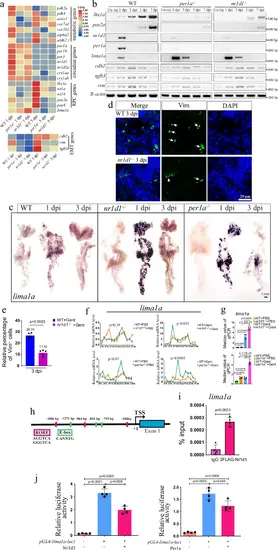
lima1a is a downstream target of the clock factorsa Heat map showing the expression of indicated genes, based on three replicates. The mesenchymal genes were placed at the bottom, and other genes were on the top. b Gel images of RT-PCR results for the indicated genes, in kidneys at the indicated time points of control, per1a−/−, and nr1d1−/− zebrafish. The RT-PCR was repeated three times, and shown were representative data. c WISH results showing the expression of lima1a at the indicated time points of control, per1a−/−, and nr1d1−/− zebrafish. WISH was repeated three times. d IF staining images of Vimentin (Vim) in 3 dpi control and nr1d1 mutant kidneys. IF experiments were repeated at least two times. e Quantification of panel d, showing the percentage of Vim+ cells. f qRT-PCR results showing the rhythmic expression of the indicated clock genes at 8 h time points over 48 h, with and without gentamicin treatment. qRT-PCR was repeated three times, and shown were representative data. g quantification of panel e. h Cartoon showing the features of lima1a gene promoter. i ChIP-PCR results showing the enrichment of Nr1d1 at lima1a promoter. The ChIP experiments were repeated two times. j Luciferase assay showing lima1a promoter activities in the presence or absence of Per1a (right) and Nr1d1 (left). The Luciferase experiments were repeated two times. p-values for each panel were indicated to show the statistical significance. ANOVA was used to test the statistical significance when multiple comparisons were conducted. |
|
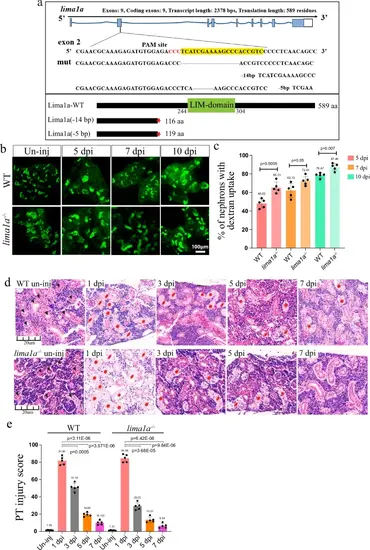
lima1a loss accelerates nephron regeneration.a CRISPR/cas9 design and the resulting in mutant alleles for lima1a. Upper: the design of CRIPSR/Cas9 and DNA sequencing results; lower: cartoon showing the full-length and respective truncated proteins. b Representative dextran fluorescent signals of kidneys at the indicated time points, in control and lima1a−/− animals. un-inj: uninjured. Green signals: dextran labeled PT. The experiments were repeated three times, and shown were representative images. c Quantification of panel b (n = 3–5 different regions of interest per group), showing the percentage of fluorescent nephrons. d HE staining images of kidneys at the indicated time points, in control and lima1a−/− animals. Black arrow heads: normal tubules, and red stars: damaged tubules. un-inj: uninjured. e Quantification of panel d (n = 4–5 different regions of interest per group), to show the renal recovery dynamics after AKI. The PT injury score is used to show the percentage of abnormal PTs. HE staining experiments were repeated three times, and shown were representative images. p-values for each panel were indicated to show the statistical significance. |
|
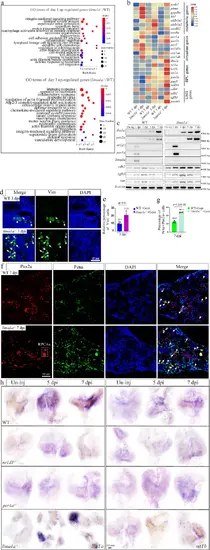
lima1a controls RPC genes via EMT.a GO terms of up-regulated genes between control and lima1a−/− kidneys at 1 and 3 dpi. b Heat map showing the expression levels of the indicated genes at the indicated time points. The RNA-seq was repeated two times, and shown were the average of the two. c Representative gel images of RT-PCR products showing the dynamic expression of the indicated genes at the indicated time points. The PCR experiments were repeated three times. d IF staining images showing the expression of Vimentin in control and lima1a−/− kidneys at the indicated time points. White arrows: Vimentin labeled cells. The experiments were repeated two times and shown were representative. e Quantification of panel d, showing the percentage of Vim+ cells. f IF staining images showing the expression of Pax2a and Pcna in control and lima1a−/− kidneys at 7 dpi. White dashed box: Pax2 expressing RPC aggregates, and white arrows: proliferating Pax2-positive cells. The experiments were repeated three times and shown to be representative. g Quantification of e (n = 5 different regions of interest per group). h Representative WISH images showing the expression of the indicated RPC marker genes at the indicated time points in control and mutant kidneys. The experiments were repeated three times and shown were from one randomly selected experiment. p-values for each panel were indicated to show the statistical significance. |