- Title
-
The conserved wobble uridine tRNA thiolase Ctu1 is required for angiogenesis and embryonic development
- Authors
- Yu, Y., Wang, C., Wang, Y., Shi, H., Hu, H., Du, Y., Zhou, Z.
- Source
- Full text @ PLoS One
|
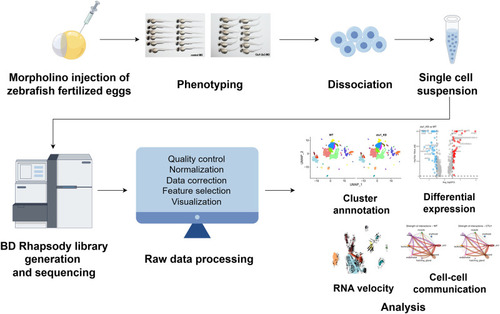
Schematic representation of workflow, figured (by Figdraw Antisense MO oligonucleotides were microinjected into fertilized one-cell stage embryos. At 2-dpf, zebrafish larvae were either photographed for phentying, or collected for single-cell preparation and sequencing using BD Rhapsody system. After filtering the raw data, further single-cell data analysis was performed such as cluster annotation, DEGs analysis, RNA velocity, cell-cell communication. |
|
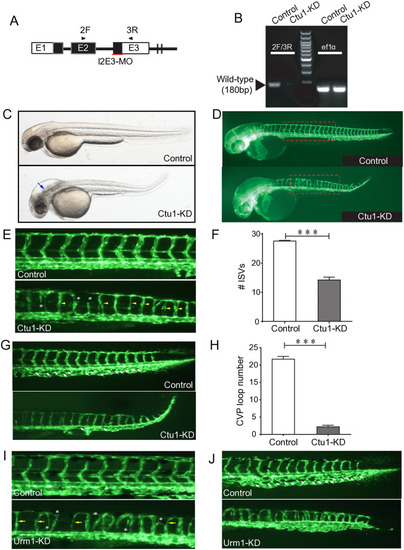
(A) Ctu1-targeted MO design strategy. (B) PCR analysis of control and Ctu1 morphant. (C and D) Bright-fieldand EGFP fluorescentimages depict the overall morphology of control and Ctu1 morphant at 2-dpf. Blue arrows indicate expanded brain ventricle and hindbrain edema in |
|
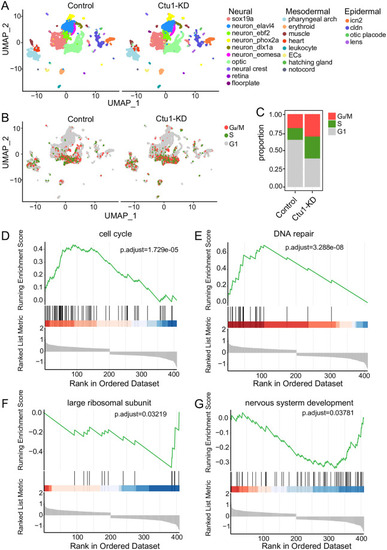
Single-cell transcriptome profiles of the UMAP visualization of all zebrafish cells, which are color-coded by cell type (A), and cell cycle phase (B). (C) The relative proportion of each cell cycle phase in the control and Ctu1 morphant. Gene sets of GSEA analysis shown are cell cycle (D), DNA repair (F), large ribosomal subunit (F) and nervous system development (G). |
|
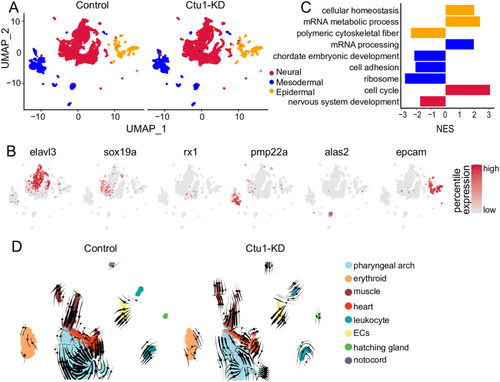
Comparative analysis of the control and (A) UMAP visualization of three germ layers. (B) UMAP visualization of key marker genes expression. Color scale represents log-normalized expression. The marker genes for neural cells are |
|
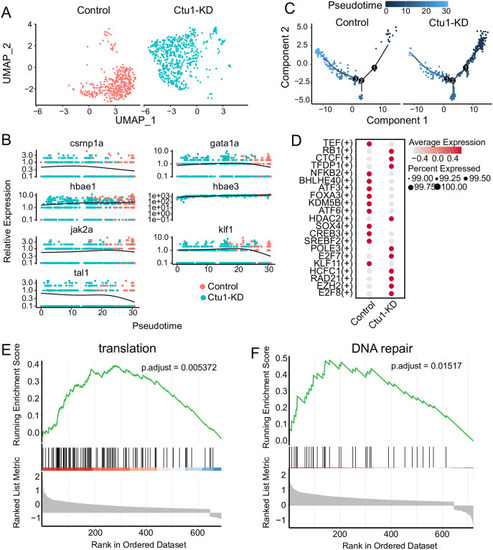
(A) UMAP visualization of erythroid cells, colored according to samples. (B) The relative expression of the marker genes associated with erythroid differentiation in the pseudotime trajectories of control and |
|
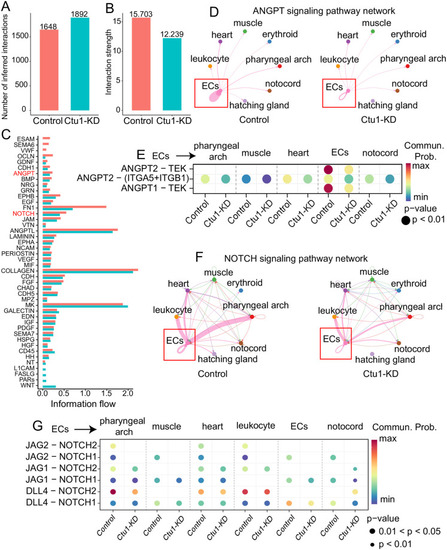
Bar plot shows overview number (A) and strength (B) in control and |
|
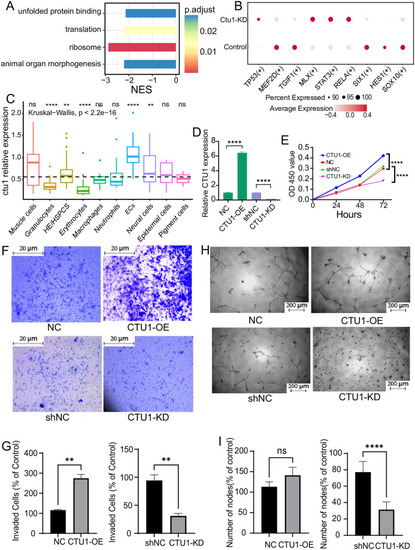
Single-cell data analysis of the endothelial cells in zebrafish, and the angiogenesis-related behaviors of human endothelial cells with differential expression of CTU1. (A) The bar plot shows signal pathways affected by |
|
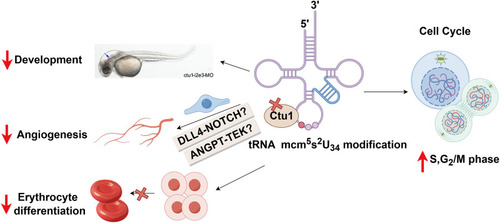
Model of the consequences of Ctu1 deficiency in zebrafish, figured by Figdraw. Recently, the essential role of mcm5s2U34 tRNA modification and their modifying enzymes in proliferation and development has been demonstrated from yeast to mammals [ |