- Title
-
Expansion of a neural crest gene signature following ectopic MYCN expression in sympathoadrenal lineage cells in vivo
- Authors
- Ibarra-García-Padilla, R., Nambiar, A., Hamre, T.A., Singleton, E.W., Uribe, R.A.
- Source
- Full text @ PLoS One
|
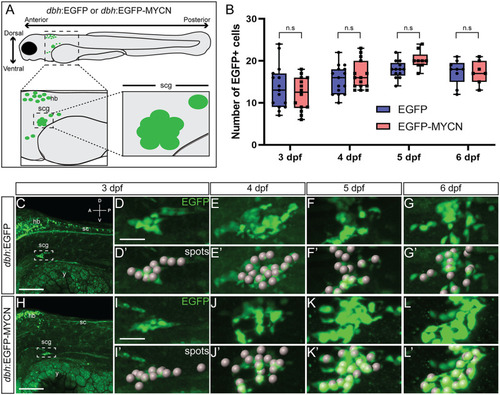
MYCN overexpression in SAP cells does not increase their cell numbers during early larval stages. A) Cartoon model of a 3 dpf |
|
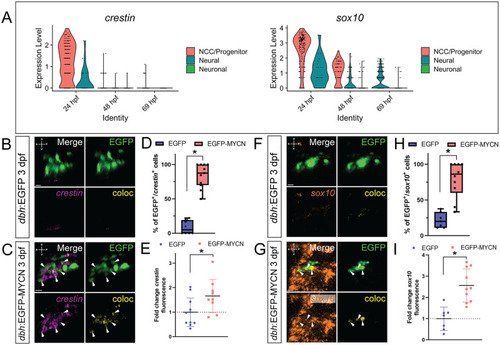
MYCN overexpressing SAP cells display an ectopic NCC gene expression signature. A) Violin plots depicting |
|
MYCN overexpression in SAP cells leads to A-F) Confocal images of the developing SCG from different time points during live imaging of |
|
MYCN overexpressing cells contain a SAP gene expression signature at 3 dpf. A,B,E,F) WICHCR performed using HCR probes against |
|
Ectopic MYCN expression results in an aberrant SAP population that co-expresses NCC and neuronal markers at 4 dpf. A) Cartoon depicting the z-slice through the SCG for quantifying individual triple positive (EGFP+/ |
|
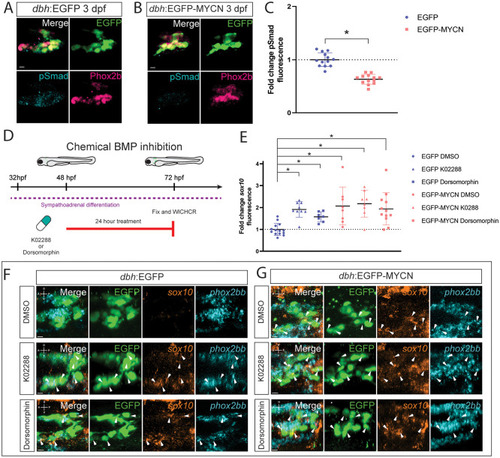
BMP signaling activity is dampened within developing EGCP-MYCN+ larvae. A,B) Representative images of SCG from |
|
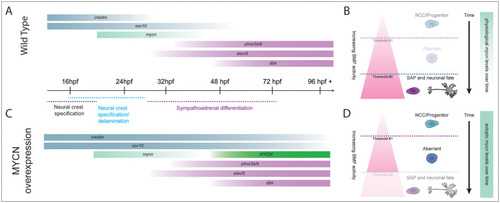
MYCN overexpression produces a cellular population with aberrant gene expression and a dampened BMP response. A,C) Normal NCC differentiation towards SAP fate requires a fine-tuned expression of transcription factors and differentiation effectors. |