- Title
-
Deciphering the function of the fifth class of Gα proteins: regulation of ionic homeostasis as unifying hypothesis
- Authors
- Abu Obaid, A., Ivandic, I., Korsching, S.I.
- Source
- Full text @ Cell. Mol. Life Sci.
|
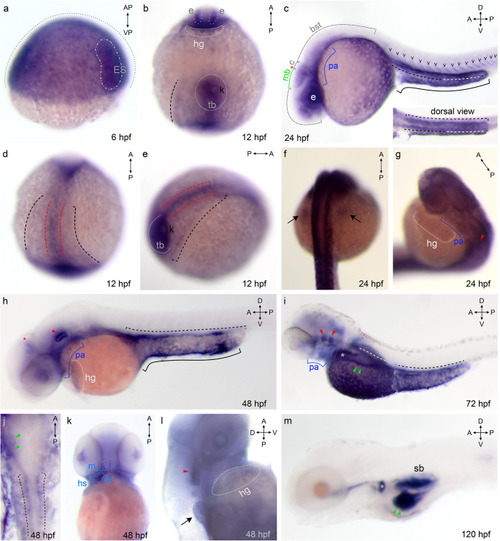
Increasingly more specific expression pattern of Gv suggests manifold roles during ontogenesis. Expression of Gv was examined by whole mount in situ hybridization in six developmental stages, 6, 12, 24, and 48 hpf (embryos), and 3 and 5 dpf (larvae). Organ abbreviations: bst, brain stem; c, cerebellum; ch, ceratohyal; e, eye; hg, hatching gland; ES, embryonic shield; hs, hyosymplectic; k, Kupffer’s vesicle; m, mandibular (Meckel’s + palatoquadrate); mb, midbrain; p.a., pharyngeal arches; sb, swim bladder; tb, tail bud. Symbols and line styles: white dotted line, hatching gland; white stippled/dotted line, hypoblast; red stippled line, notochord (axial mesoderm at 12 hpf); red asterisk, midbrain/hindbrain boundary; red arrow head, inner ear; green line, midbrain; green arrow head, gut; cyan stippled line, first and second pharyngeal arches (mandibular: Meckel’s cartilage and palatoquadrate and hyoid: ceratohyal and hyosymplectic); blue line, pharyngeal arches; gray dotted line, epiblast; gray asterisk, proximal convoluted tubule (PCT); black stippled line, pronephros (lateral plate mesoderm at 12 hpf); black line, yolk extension; black arrow, pectoral fin bud; black ‘v’, somites. For better visibility some lines are contrast adapted. EXPRESSION / LABELING:
|
|
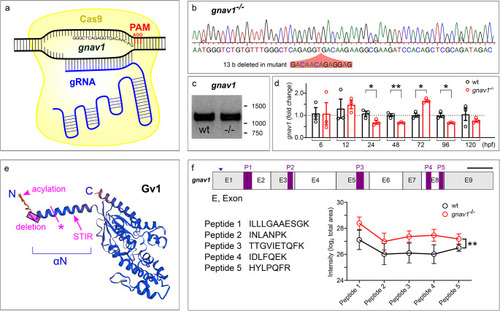
Knockout of zebrafish gnav1. ( |
|
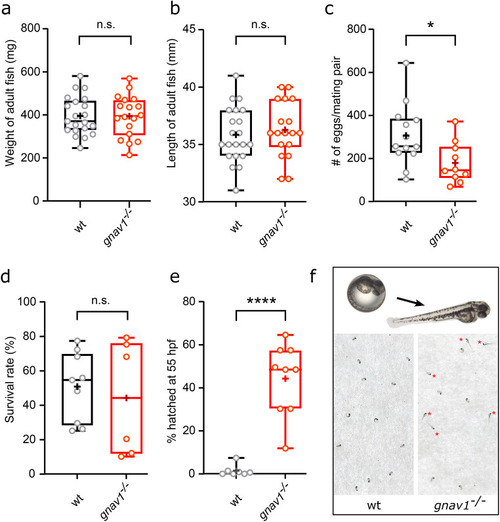
Reduced oviposition and premature hatching in Gv knockout. Wildtype, wt; gnav1 deletion mutant, gnav1−/−. Data for panels a) to e) are shown as whisker plots with quartile segments, mean values are indicated by crosses, individual measurements are shown as circles. ( |
|
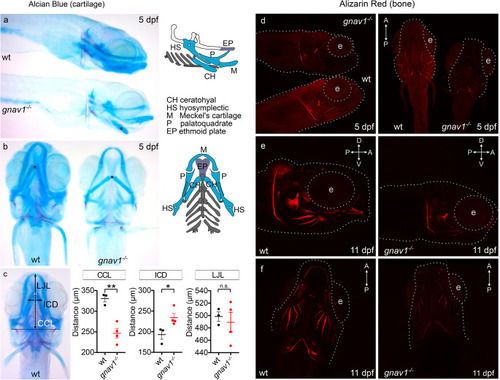
Craniofacial abnormalities in the |
|
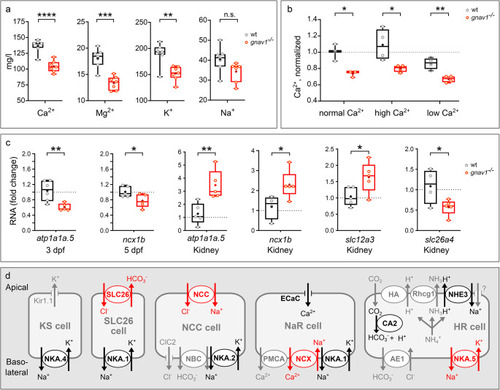
Decreased cation levels and altered ion transporter gene expression in Gv mutant zebrafish. Data for panels EXPRESSION / LABELING:
PHENOTYPE:
|
|
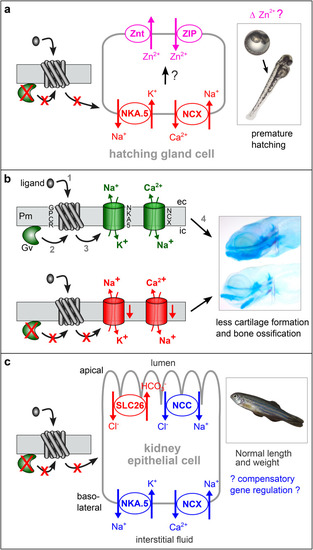
Hypotheses for Gv signalling pathways in three developmental stages. Arrows may represent several molecular steps. Red color indicates block (red crosses) or decrease in the mutant, blue depicts an increase, green represents wildtype levels, magenta shows hypothetical effect. ( |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|

Unillustrated author statements |