- Title
-
Microglia Mitigate Neuronal Activation in a Zebrafish Model of Dravet Syndrome
- Authors
- Brenet, A., Somkhit, J., Csaba, Z., Ciura, S., Kabashi, E., Yanicostas, C., Soussi-Yanicostas, N.
- Source
- Full text @ Cells
|
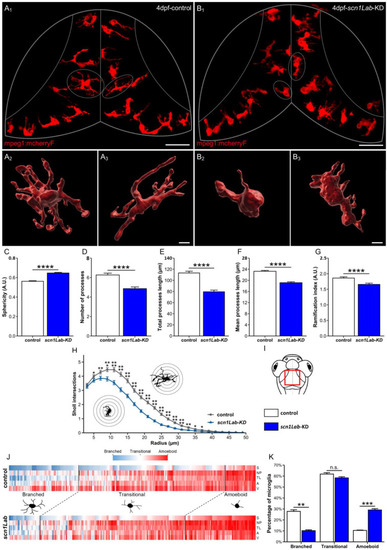
Microglia morphology parameters and clustering, in |
|
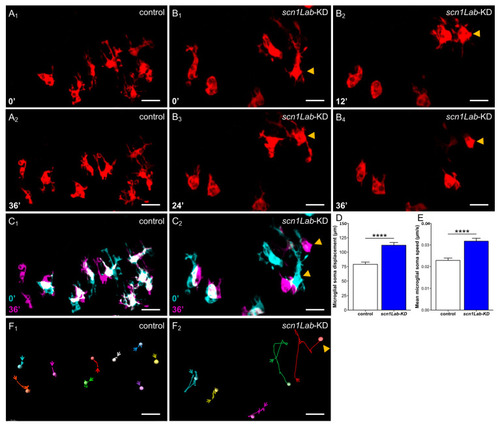
Microglia dynamics in |
|
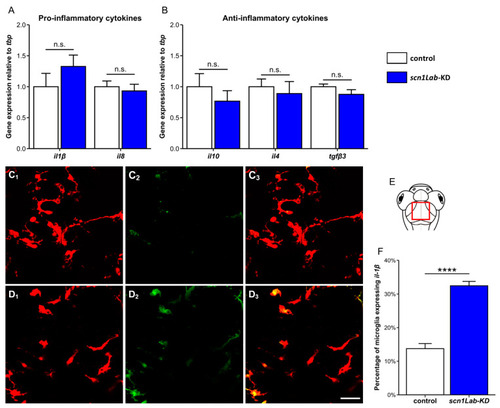
Cytokine expression in |
|
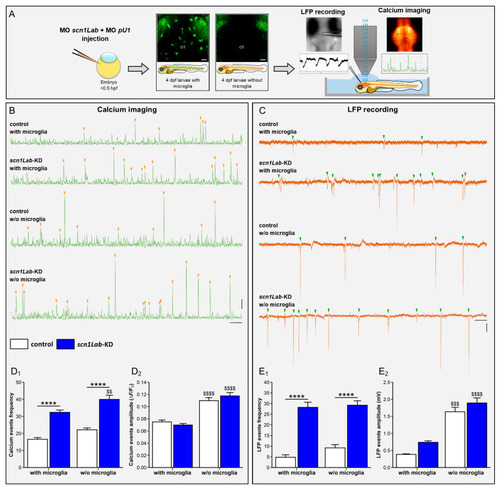
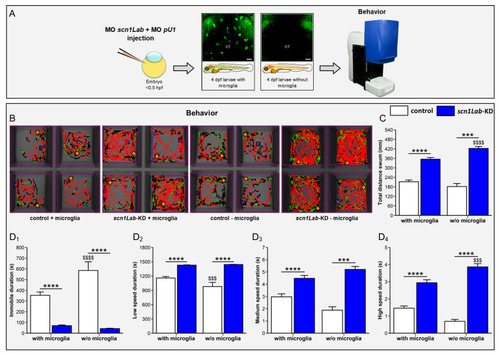
Microglia depletion increases neuron hyperactivity in |
|
Microglia ablation increases the locomotor activity of |