- Title
-
Kalium channelrhodopsins effectively inhibit neurons
- Authors
- Ott, S., Xu, S., Lee, N., Hong, I., Anns, J., Suresh, D.D., Zhang, Z., Zhang, X., Harion, R., Ye, W., Chandramouli, V., Jesuthasan, S., Saheki, Y., Claridge-Chang, A.
- Source
- Full text @ Nat. Commun.
|
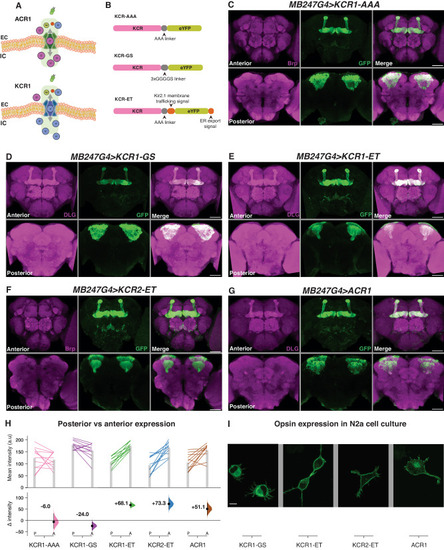
Membrane-trafficking signals improve KCR localization to axons. |
|
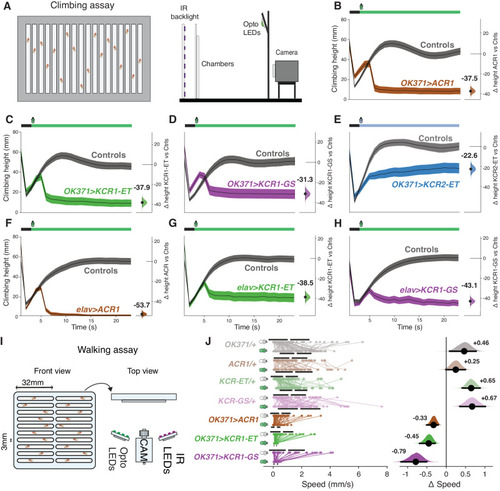
KCR actuation inhibits climbing and walking in |
|
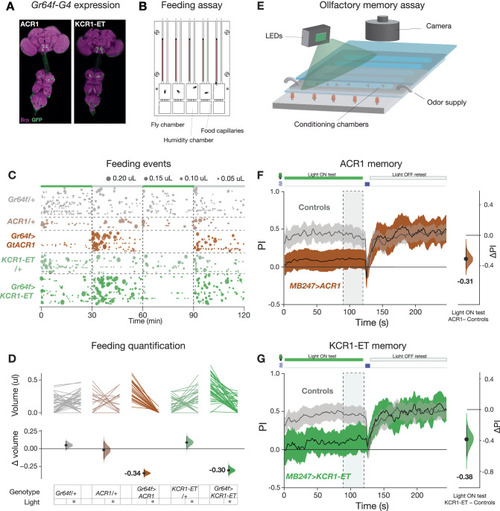
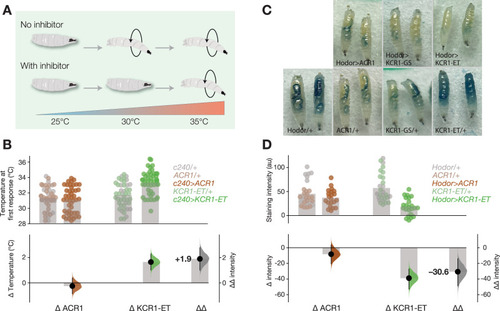
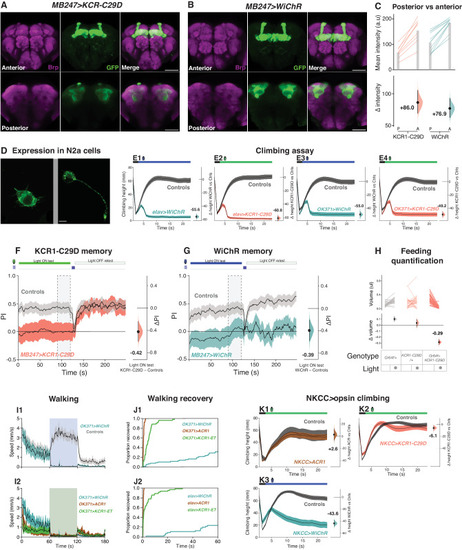
KCR1 and ACR1 actuation show comparable effects on feeding and memory. |
|
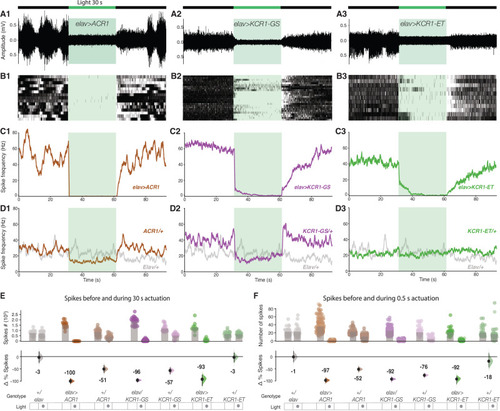
Spontaneous spiking in a larval abdominal nerve is silenced by KCR1. |
|
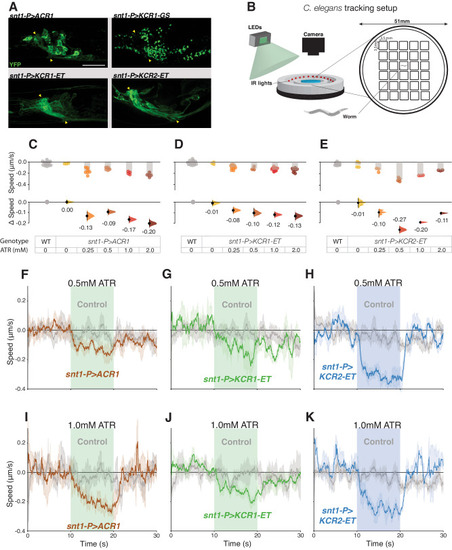
KCR actuation impairs locomotion in |
|
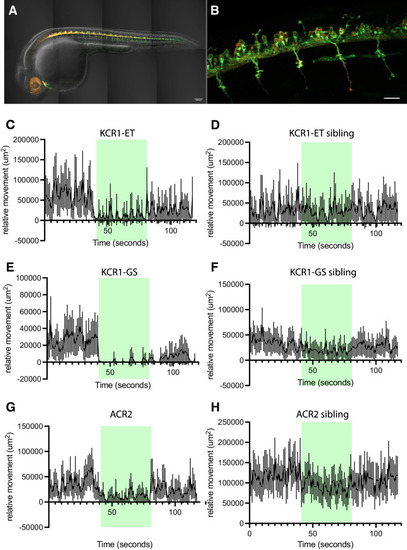
KCR1 actuation inhibits zebrafish larval movements. |
|
KCR1-ET affects cells with non-canonical chloride signaling. |
|
KCRs with improved K+ selectivity have increased potency. |