|
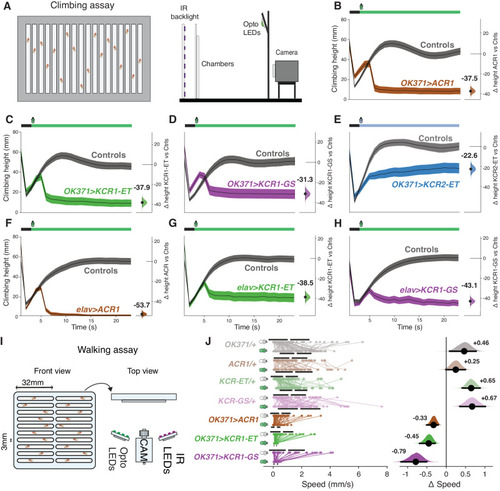
KCR actuation inhibits climbing and walking in Drosophila. A Schematic of the single-fly climbing assay, showing the chamber on the left and the different assay elements on the right. B–E Averaged climbing performance of flies expressing the respective opsin with OK371-Gal4 and the corresponding Gal4 driver and UAS responder controls (gray) in the presence of light. In the schematic bar (top), black indicates the 3s baseline, and the colored bar indicates the illumination interval. The y-axis on the right indicates the mean difference in effect size between the genotypic controls and test flies. The last 10s of the experiment were used for effect size comparisons. Error bands represent the 95% CI. OK371 > ACR1 test, n = 136 biologically independent animals over 8 independent experiments, OK371 > ACR1 controls, n = 255 biologically independent animals over 15 independent experiments. OK371 > KCR1-ET test, n = 119 biologically independent animals over 8 independent experiments, OK371 > KCR1-ET controls, n = 238 biologically independent animals over 14 independent experiments. OK371 > KCR1-GS test, n = 136 biologically independent animals over 8 independent experiments, OK371 > KCR1-GS controls, n = 221 biologically independent animals over 13 independent experiments. OK371 > KCR2-ET test, n = 116 biologically independent animals over 7 independent experiments, OK371 > ACR1 controls, n = 136 biologically independent animals over 8 independent experiments. F–H Averaged climbing performance of flies expressing the respective opsin with elav-Gal4 and the corresponding genotypic controls in the presence of light. Green illumination intensity was 11 μW/mm2. The blue illumination intensity of KCR2-ET was 85 μW/mm2. Error bands represent the 95% CI. elav > ACR1 test, n = 153 biologically independent animals over 9 independent experiments, elav > ACR1 controls, n = 255 biologically independent animals over 15 independent experiments. elav > KCR1-ET test, n = 135 biologically independent animals over 8 independent experiments, elav > KCR1-ET controls, n = 255 biologically independent animals over 15 independent experiments. elav > KCR1-GS test, n = 117 biologically independent animals over 7 independent experiments, elav > KCR1-GS controls, n = 255 biologically independent animals over 15 independent experiments. I Schematic representation of the walking assay with the chamber view from the front (left) and the experimental setup view from the top (right). J Walking-speed comparisons before and during illumination (24 µW/mm2). The left side of the plot displays the activity of individual flies (dots), and the gap between the horizontal error bars represents the mean. The right side of the plot displays the speed mean difference effect size for each respective genotype. Error bars represent the 95% CI. OK371/+,n = 75 biologically independent animals over 3 independent experiments. ACR1/+,n = 61 biologically independent animals over 3 independent experiments. KCR1-ET/+,n = 63 biologically independent animals over 3 independent experiments. KCR1-GS/+,n = 73 biologically independent animals over 3 independent experiments. OK371 > ACR1,n = 69 biologically independent animals over 3 independent experiments. OK371 > KCR1-ET,n = 57 biologically independent animals over 3 independent experiments. OK371 > KCR1-GS,n = 48 biologically independent animals over 2 independent experiments. Additional statistical information for all panels is presented in Supplementary Dataset 1. Source data are provided as a Source Data file.
|