- Title
-
The transcription of the main gene associated with Treacher-Collins syndrome (TCOF1) is regulated by G-quadruplexes and cellular nucleic acid binding protein (CNBP)
- Authors
- Gil Rosas, M., Centola, C., Torres, M., Mouguelar, V.S., David, A.P., Piga, E.J., Gomez, D., Calcaterra, N.B., Armas, P., Coux, G.
- Source
- Full text @ Sci. Rep.
|
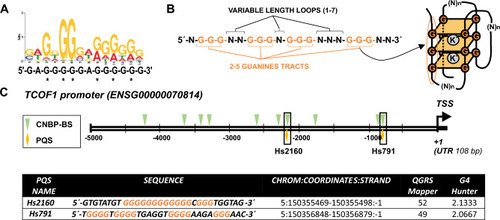
CNBP binding sites and PQSs detected in |
|
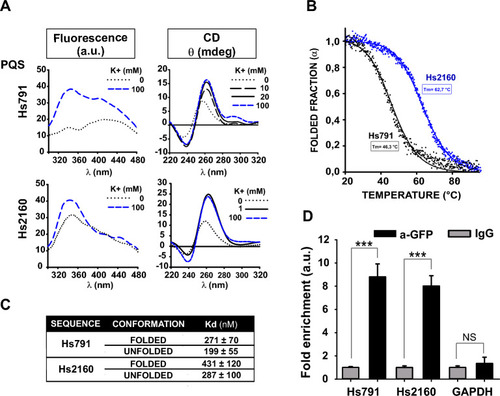
In vitro G4 folding assessment and CNBP binding of the Hs791 and Hs2160 PQSs. ( |
|
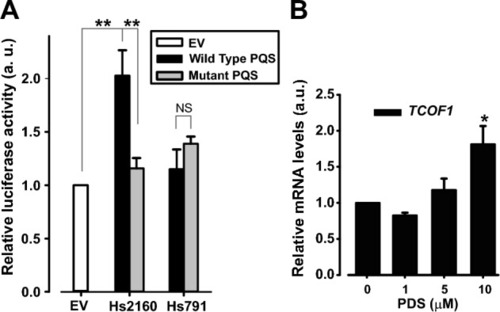
Role of Hs791 and Hs2160 on transcriptional expression control. ( |
|
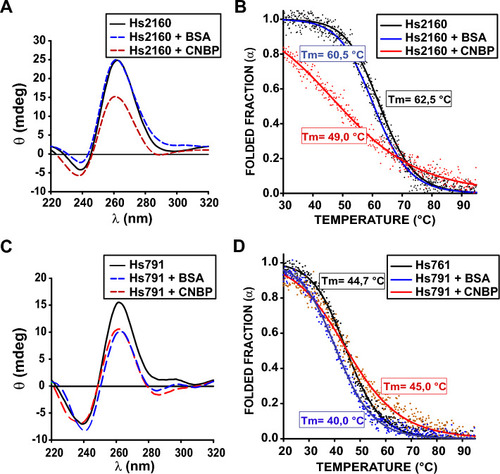
In vitro CNBP action on folded Hs791 and Hs2160. ( |
|
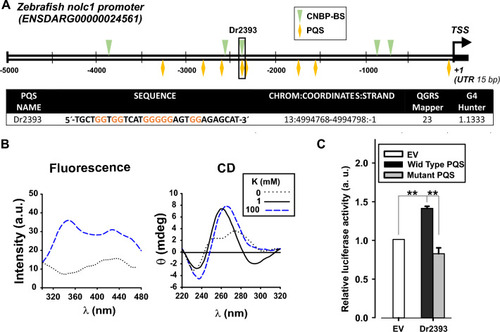
Promoter sequence analysis of the |
|
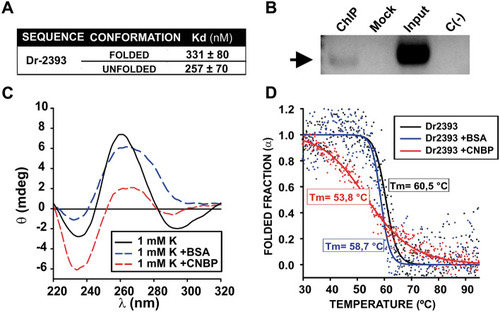
CNBP binding and action over Dr2393. ( |
|
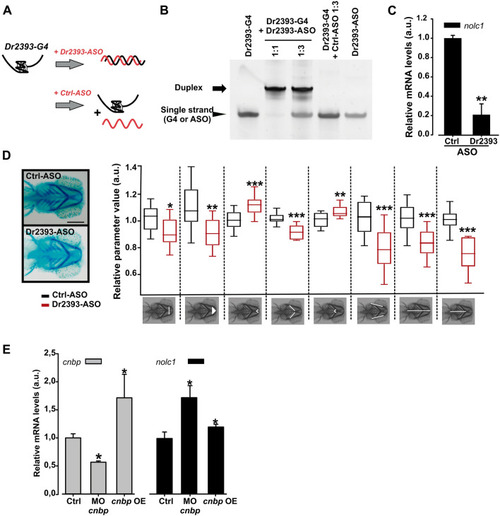
Effect of Dr2393 disruption and CNBP varying levels on zebrafish |
|
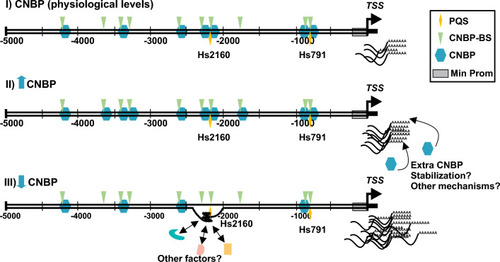
Diagram depicting the working model of |