- Title
-
Telomerase RNA-based aptamers restore defective myelopoiesis in congenital neutropenic syndromes
- Authors
- Martínez-Balsalobre, E., García-Castillo, J., García-Moreno, D., Naranjo-Sánchez, E., Fernández-Lajarín, M., Blasco, M.A., Alcaraz-Pérez, F., Mulero, V., Cayuela, M.L.
- Source
- Full text @ Nat. Commun.
|
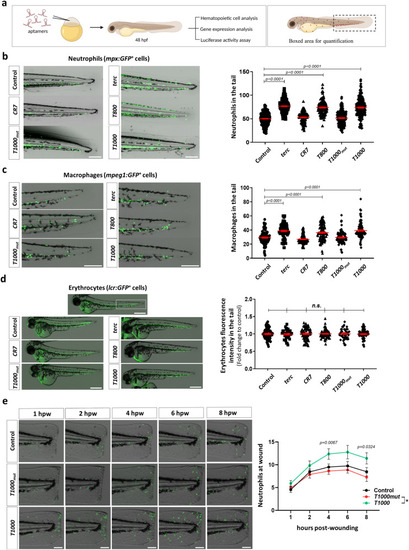
T800 and T1000 aptamers enforce myelopoiesis in zebrafish. |
|
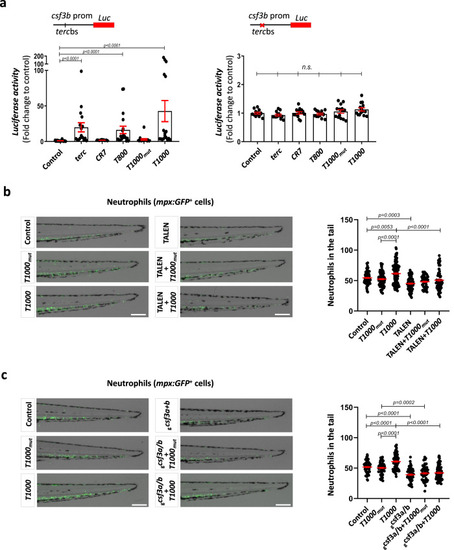
T1000 aptamer enforce myelopoiesis by activating |
|
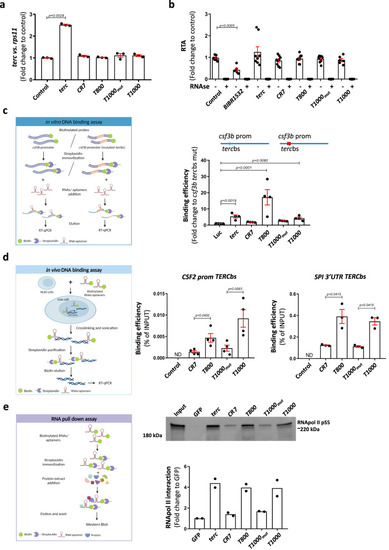
Aptamers physically bind to DNA and to RNApolII. |
|
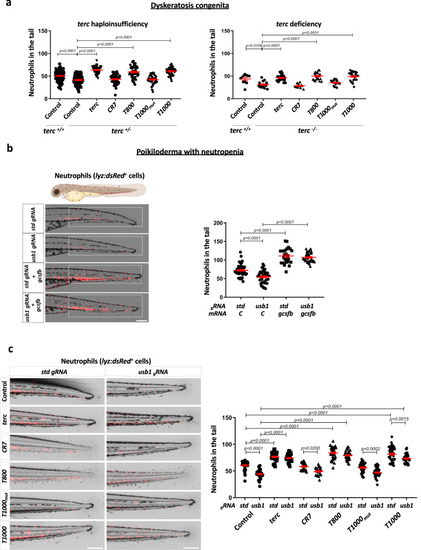
Aptamers rescue neutropenia in zebrafish models of DC and PN. |
|
Aptamers rescue defective myelopoiesis in mouse and iPSC from DC patients. |