- Title
-
Actionable cancer vulnerability due to translational arrest, p53 aggregation and ribosome biogenesis stress evoked by the disulfiram metabolite CuET
- Authors
- Kanellis, D.C., Zisi, A., Skrott, Z., Lemmens, B., Espinoza, J.A., Kosar, M., Björkman, A., Li, X., Arampatzis, S., Bartkova, J., Andújar-Sánchez, M., Fernandez-Capetillo, O., Mistrik, M., Lindström, M.S., Bartek, J.
- Source
- Full text @ Cell Death Differ.
|
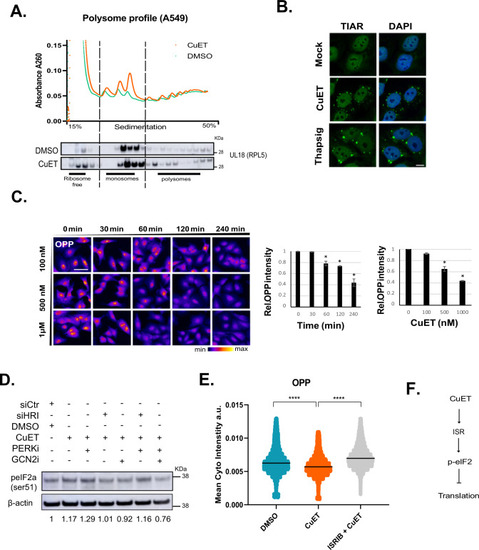
CuET rapidly blocks protein synthesis. |
|
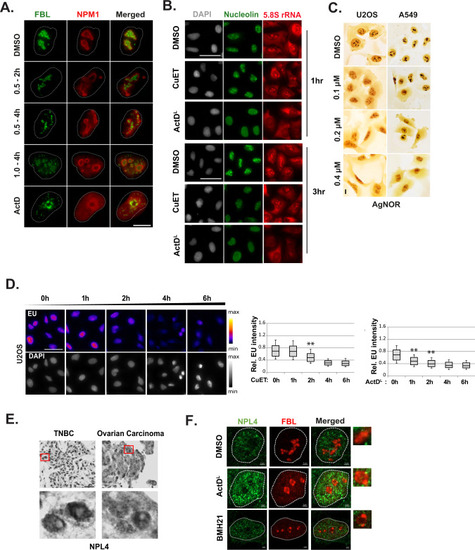
CuET alters the nucleolar morphology. |
|
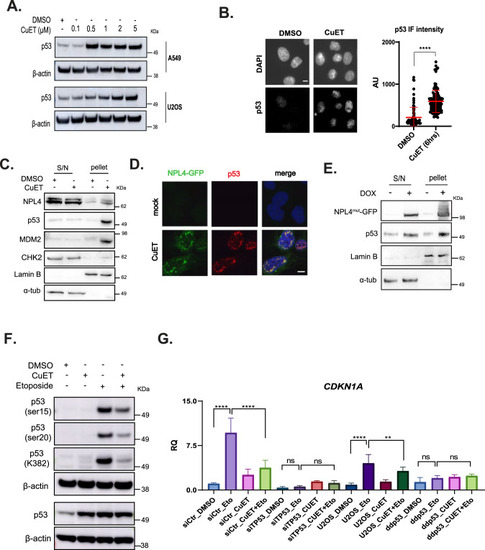
CuET triggers p53 entrapment in NPL4-rich aggregates. |
|
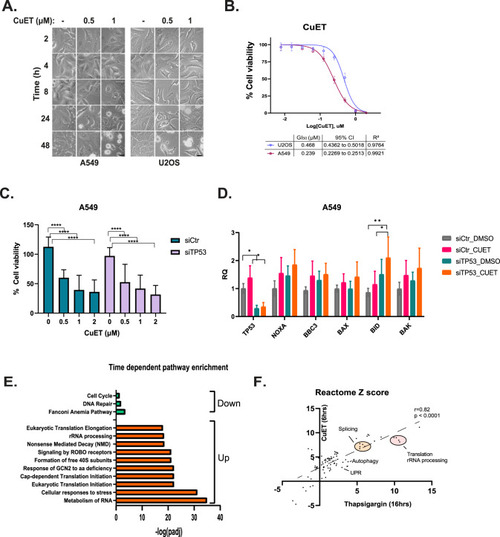
CuET induces cell death in a p53-independent manner. |
|
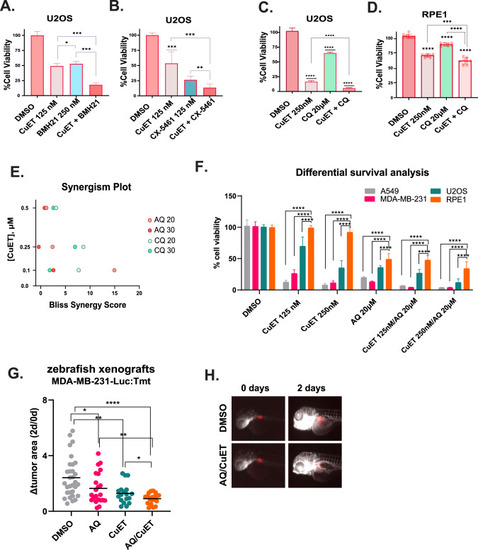
RiBi and autophagy inhibition potentiate the cytotoxic effect of CuET. |
|
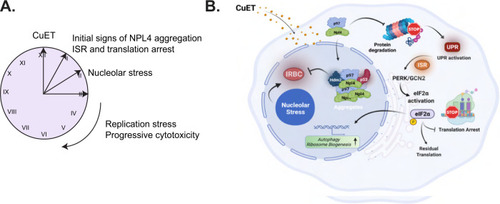
Schematic illustration of the proposed model. |