- Title
-
The microbiota regulates hematopoietic stem and progenitor cell development by mediating inflammatory signals in the niche
- Authors
- Zhong, D., Jiang, H., Zhou, C., Ahmed, A., Li, H., Wei, X., Lian, Q., Tastemel, M., Xin, H., Ge, M., Zhang, C., Jing, L.
- Source
- Full text @ Cell Rep.
|
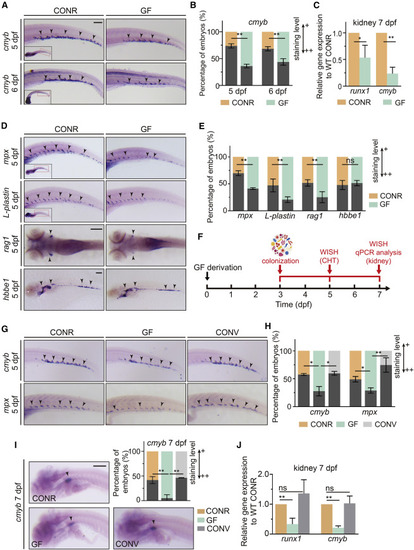
The commensal microbiota is required for HSPC development (A) CONR and GF zebrafish were examined by WISH for cmyb expression in the caudal hematopoietic tissue (CHT) region at 5 and 6 dpf. Arrowheads denote cmyb+ HSPCs. (B) Quantification of WISH results in (A). (C) qPCR analysis of runx1 and cmyb expression in the kidneys of CONR and GF zebrafish at 7 dpf. (D) WISH for neutrophil marker (mpx), leukocyte marker (L-plastin), T lymphocyte marker (rag1), and erythrocyte marker (hbbe1) at 5 dpf in CONR and GF zebrafish. (E) Quantification of WISH results in (D). (F) The experimental timeline for the conventionalization (CONV) experiments in (G–J). (G) WISH for cmyb and mpx in CONR, GF, and CONV fish in the CHT region at 5 dpf. (H) Quantification of WISH results in (G). (I) WISH for cmyb in the kidney in CONR, GF, and CONV fish at 7 dpf. (J) qPCR analysis of runx1 and cmyb expression in the kidneys of CONR, GF, and CONV zebrafish at 7 dpf. Student’s t tests in (B, C, and E), ANOVA in (H, I, and J); mean ± SEM from more than three independent experiments, n = 45–60 larvae per group for WISH, 30–40 larvae per group for qPCR, ∗p < 0.05, ∗∗p < 0.01; ns, not significant. CONR, conventionally raised; GF, germ free; CONV, conventionalized. Scale bars, 200 μm. See also Figures S1 and S2. |
|
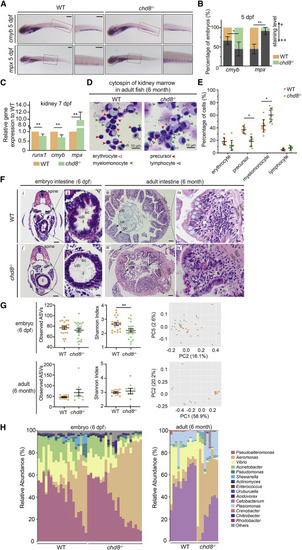
chd8−/− zebrafish display inhibited HSPC formation, impaired intestinal structure, and altered microbiota in larva and adults (A) WISH for cmyb and mpx in the CHT in WT and chd8−/− fish at 5 dpf. (B) Quantification of WISH results from (A). Scale bars, 200 μm. (C) qPCR analysis for runx1, cmyb and mpx expression in the kidneys of WT and chd8−/− fish at 7 dpf. (D) May-Grunwald-Giemsa staining of the whole kidney marrow cells obtained from 6-month-old WT and chd8−/− fish. Open red arrows indicate erythroid cells. Red arrows indicate precursors. Green arrows indicate myelomonocytes. Open black arrows indicate lymphoid cells. (E) The statistical analysis for blood cell counts, n = 6 in WT and n = 5 in chd8−/−. (F) Representative images of intestinal cross-sections of WT and chd8−/− larval and adult fish, stained with hematoxylin and eosin. ec, enterocytes; il, intestinal lumen; ce, columnar epithelium; gc, goblet cells,; lsm, longitudinal smooth muscle layer; csm, circular smooth muscle layer. Scale bars, 50 μm (i, i’, iii, and iii’) and 10 μm (ii, ii’ iv, and iv’). (G) The analysis of alpha diversity for microbiota in WT and chd8−/− larvae and adults. The PCoA plots based on Bray-Curtis distance for the microbial structure of WT and chd8−/− larvae (p = 0.007, PERMANOVA test based on Bray-Curtis distance) and adults (p = 0.001, PERMANOVA test based on Bray-Curtis distance). Orange, WT; green, chd8−/−. (H) Relative abundance of the main genera of microbiota in WT and chd8−/− larvae and adults. n = 20 in WT and n = 22 in chd8−/ larvae; n = 11 in WT and n = 9 in chd8−/ adults. In (B), (C), and (E), Student’s t tests; mean ± SEM of three independent experiments (B and C) or of all samples (G), n = 45–60 larvae in each group for WISH, n = 30–40 larvae per group for qPCR, ∗p < 0.05, ∗∗p < 0.01; ns, not significant. See also Figure S3. |
|
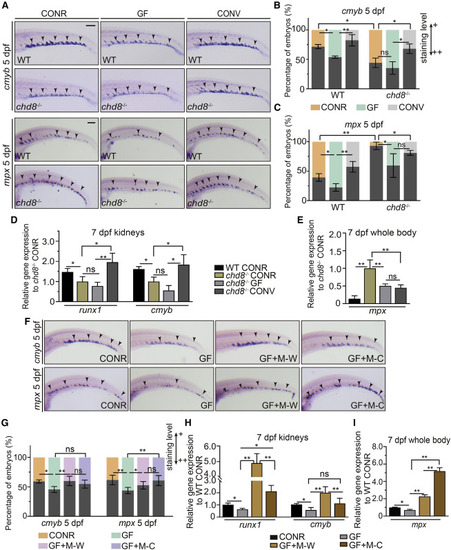
Dysregulated microbiota in chd8−/− inhibits HSPC development and enhances myeloid differentiation (A) WISH for cmyb and mpx in 5-dpf WT and chd8−/− CONR and GF larvae, and in WT and chd8−/− GF larvae at 3 dpf conventionalized with microbiota from the CONR WT (CONV). (B and C) Quantification of WISH results from (A). (D) qPCR analysis of runx1 and cmyb expression in the kidneys of WT CONR, chd8−/− CONR, GF, and CONV larvae at 7 dpf. (E) qPCR analysis of mpx expression in the whole body of WT CONR, chd8−/− CONR, GF, and CONV zebrafish at 7 dpf. (F) WISH for cmyb and mpx in 5-dpf WT CONR, GF larvae, and GF larvae at 3 dpf conventionalized with WT-sourced microbiota (M-W) or chd8−/−-sourced microbiota (M-C) obtained from adult intestines. (G) Quantification of WISH results from (F). (H) qPCR analysis of runx1 and cmyb expression in the kidneys at 7 dpf of WT CONR, GF, and GF larvae at 3 dpf conventionalized with M-W or M-C. (I) The mpx expression in the whole body at 7 dpf of WT CONR, GF, and GF larvae at 3 dpf conventionalized with M-W or M-C. ANOVA in all analyses; mean ± SEM of three independent experiments, n = 45–60 larvae per group for WISH, 30–40 larvae per group for qPCR analysis, ∗p < 0.05, ∗∗p < 0.01, ns, not significant. Scale bars, 200 μm. |
|
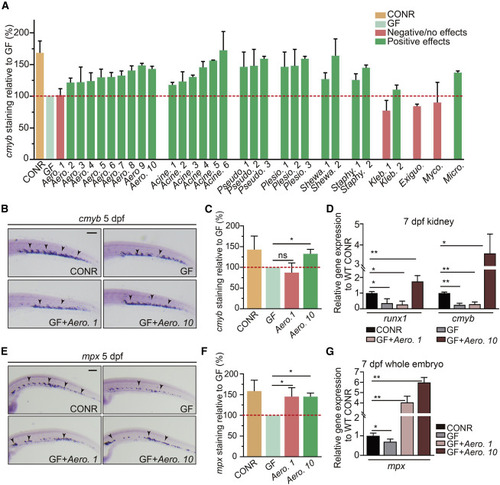
Individual bacterial members of zebrafish microbiota exhibit differential effects on HSPC development (A) Quantification of cmyb staining in WT CONR, GF larvae, and GF larvae at 3 dpf mono-associated with a specific bacterial strain. Bacterial mono-associations are labeled by genus. Different bacterial species within the same genus are labeled with a number (1, 2, 3, and so on). (B) WISH for cmyb in WT CONR, GF larvae, and GF larvae treated with bacterial strains Aero. 1 or Aero. 10. (C) Quantification of WISH results from (B). (D) qPCR analysis of runx1 and cmyb expression in the kidney at 7 dpf of WT CONR, GF larvae, and GF larvae treated with Aero. 1 or Aero. 10. (E) WISH for mpx in WT CONR, GF larvae, and GF larvae treated with Aero. 1 or Aero. 10. (F) Quantification of WISH results from (E). (G) qPCR analysis of mpx expression at 7 dpf in whole embryos of WT CONR, GF larvae, and GF larvae treated with Aero. 1 or Aero. 10. ANOVA in all analyses; mean ± SEM of three independent experiments, n = 45–60 larvae per group for WISH, 30–40 larvae per group for qPCR analysis, ∗p < 0.05, ∗∗p < 0.01, ns, not significant. Scale bars, 200 μm. See also Figure S4. |
|
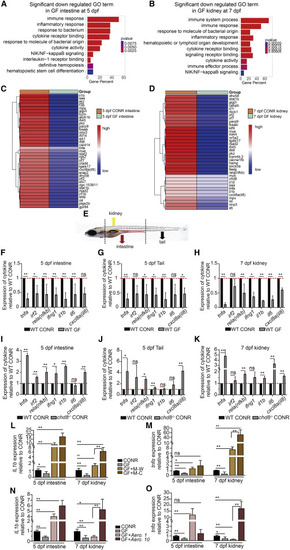
Microbiota mediates inflammatory signals in the intestine, tail, and kidney (A and B) Gene ontology (GO) analysis of differentially expressed genes revealed inflammatory signaling enriched in downregulated genes in the intestine (A) and kidney (B) after GF treatment. (C and D) Heatmap analysis showed comparison of gene expression in the intestine (C) and kidney (D) between CONR and GF larvae. Multiple inflammatory signaling genes were decreased in GF larvae. (E) Schematic representation of tissues collected from the intestine (5 dpf), tail (including CHT region, 5 dpf), and kidney (7 dpf) for qPCR analysis. (F–H) qPCR analysis of representative cytokine genes in the intestine, tail, and kidney in WT CONR and GF larvae. (I–K) qPCR analysis of cytokine genes in the intestine, tail, and kidney in CONR WT and chd8−/− larvae. (L and M) qPCR analysis of il1b (L) and tnfa (M) in the intestine and kidney obtained from WT CONR, GF larvae, and GF larvae at 3 dpf treated with WT-sourced microbiota (M-W) or chd8−/−-sourced microbiota (M-C). (N and O) qPCR analysis of il1b (N) and tnfa (O) in the intestine and kidney obtained from WT CONR, GF larvae, and GF larvae treated with Aero. 1 or Aero. 10. Student’s t tests in (F–K), ANOVA in (L–O); mean ± SEM of more than three independent experiments, n = 30–40 larvae per group for qPCR analysis, ∗p < 0.05, ∗∗p < 0.01; ns, not significant. See also Figures S5 and S6. |
|
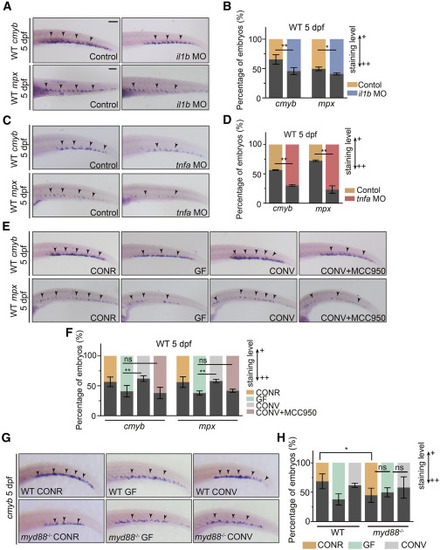
Microbiota-promoted HSPC development is dependent on basal inflammatory cytokine expression (A) WISH for cmyb and mpx in control and il1b morphant larvae. (B) Quantification of WISH results from (A). (C) WISH for cmyb and mpx in control and tnfa morphant larvae. (D) Quantification of WISH results from (C). (E) WISH for cmyb and mpx in WT CONR, GF larvae, and GF larvae at 3 dpf treated with microbiota from the CONR WT (CONV) with or without MCC950. (F) Quantification of WISH results from (E). (G) WISH for cmyb in WT and myd88−/− CONR, GF larvae, and GF larvae at 3 dpf treated with microbiota from the CONR WT (CONV). (H) Quantification of WISH results from (G). Student’s t tests in (B and D), ANOVA in (F and H); means ± SEM of three independent experiments, n = 45–60 larvae per group for WISH, ∗p < 0.05, ∗∗p < 0.01; ns, not significant. Scale bars, 200 μm. See also Figure S7. |
|
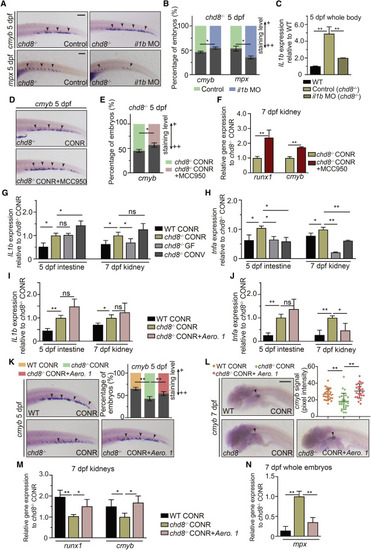
Selective inhibition of elevated cytokine expression in chd8−/− kidney by microbiota intervention rebalances HSPC development and myeloid differentiation (A) WISH for cmyb and mpx in chd8−/− and chd8−/− larvae injected with il1b MO. (B) Quantification of WISH results from (A). (C) qPCR analysis of il1b expression at 5 dpf in the whole embryos of WT, chd8−/−, and chd8−/− larvae injected with il1b MO. (D) WISH for cmyb at 5 dpf in chd8−/− and chd8−/− larvae treated with MCC950 from 3 dpf. (E) Quantification of WISH results from (D). (F) qPCR analysis of runx1 and cmyb expression in kidney at 7 dpf of chd8−/− CONR and chd8−/− CONR larvae treated with MCC950 from 3 dpf. (G and H) qPCR analysis of il1b (G) and tnfa (H) in the intestine and kidney obtained from WT CONR, chd8−/− CONR, chd8−/− GF larvae, and chd8−/− GF larvae treated with microbiota from the CONR WT at 3dpf. (I and J) qPCR analysis of il1b (I) and tnfa (J) in the intestine and kidney obtained from WT CONR, chd8−/− CONR larvae, and chd8−/− CONR larvae treated with Aero. 1 at 3 dpf. (K) WISH for cmyb at 5 dpf in WT CONR, chd8−/− CONR, and chd8−/− CONR larvae treated with Aero. 1 at 3 dpf. (L) WISH for cmyb in the kidney at 7 dpf in WT CONR, chd8−/− CONR, and chd8−/− CONR larvae treated with Aero. 1 at 3 dpf, each data point represents an individual larva, n = 26–30 per group. (M) qPCR analysis of runx1 and cmyb expression in the kidney of WT CONR, chd8−/− CONR larvae, and chd8−/− CONR larvae treated with Aero. 1 at 3 dpf. (N) qPCR analysis of mpx expression in the whole embryos in WT CONR, chd8−/− CONR larvae, and chd8−/− CONR larvae treated with Aero. 1 at 3 dpf. Student’s t tests in (B, C, E, and F), ANOVA in (G–J and L–N); means ± SEM of three independent experiments, n = 45–60 larvae per group for WISH, n = 30–40 larvae per group for qPCR, ∗p < 0.05, ∗∗p < 0.01; ns, not significant. Scale bars, 200 μm. See also Figure S7. |