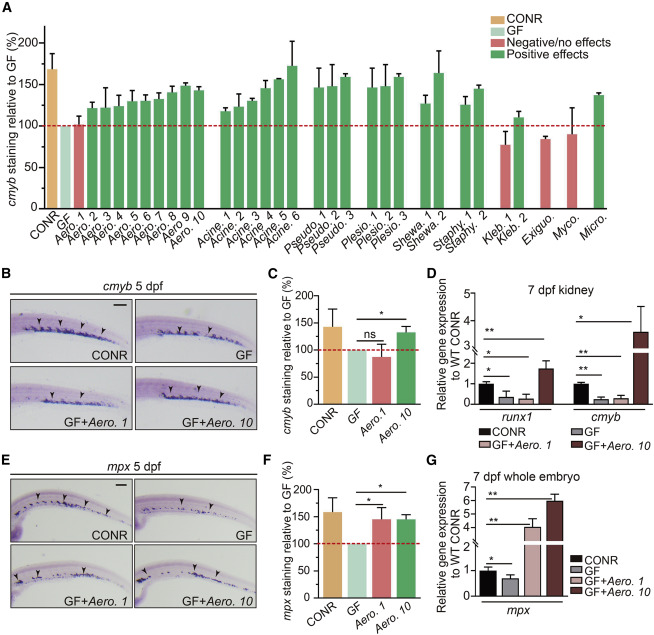
Fig. 4 Individual bacterial members of zebrafish microbiota exhibit differential effects on HSPC development (A) Quantification of cmyb staining in WT CONR, GF larvae, and GF larvae at 3 dpf mono-associated with a specific bacterial strain. Bacterial mono-associations are labeled by genus. Different bacterial species within the same genus are labeled with a number (1, 2, 3, and so on). (B) WISH for cmyb in WT CONR, GF larvae, and GF larvae treated with bacterial strains Aero. 1 or Aero. 10. (C) Quantification of WISH results from (B). (D) qPCR analysis of runx1 and cmyb expression in the kidney at 7 dpf of WT CONR, GF larvae, and GF larvae treated with Aero. 1 or Aero. 10. (E) WISH for mpx in WT CONR, GF larvae, and GF larvae treated with Aero. 1 or Aero. 10. (F) Quantification of WISH results from (E). (G) qPCR analysis of mpx expression at 7 dpf in whole embryos of WT CONR, GF larvae, and GF larvae treated with Aero. 1 or Aero. 10. ANOVA in all analyses; mean ± SEM of three independent experiments, n = 45–60 larvae per group for WISH, 30–40 larvae per group for qPCR analysis, ∗p < 0.05, ∗∗p < 0.01, ns, not significant. Scale bars, 200 μm. See also Figure S4.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Cell Rep.