- Title
-
Stage-specific and cell type-specific requirements of ikzf1 during haematopoietic differentiation in zebrafish
- Authors
- Hess, I., Sagar, ., O Meara, C., Grün, D., Schorpp, M., Boehm, T.
- Source
- Full text @ Sci. Rep.
|
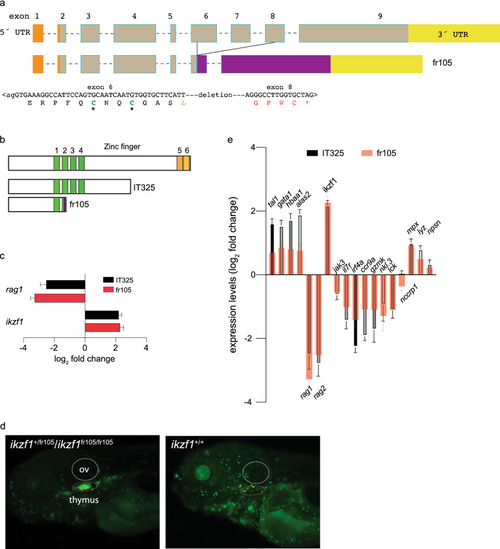
Characterization of a new ikzf1 allele. (a) Schematic structure of the zebrafish ikzf1 gene. Exons are represented by boxes and numbered and drawn to scale; the introns are indicated by dashed lines but are not drawn to scale. The 5´-UTR and 3´-UTR are indicated in orange and yellow. The CRISPR/Cas9-induced deletion in the fr105 allele is indicated; the nucleotide sequence across the junction is shown and the resulting protein sequence is given in single letter code. The deletion creates a frame-shift, resulting in the addition of unrelated amino acid residues (red letters) before a premature stop codon (*). (b) Schematic of the proteins encoded by the two ikzf1 alleles studied here; the IT325 allele was described by Schorpp et al.9. (c) Lack of larval T cell development in homozygous IT325 and fr105 mutants as determined by RNA-seq at 5 dpf; rag1 and ikzf1 expression levels relative to wildtype are shown (mean ± s.e.m.; n = 3 biological replicas). (d) Analysis of the ikzf1fr105 allele. In the ikaros:eGFP background, wild-type and heterozygous fish cannot be phenotypically distinguished (left panel), whereas the mutants (right panel) lack fluorescent signals in the thymus (dashed red oval), and exhibit brighter fluorescence. ov, otic vesicle. e Differential gene expression patterns in IT325 and fr105 mutants at 5 dpf. The log2 fold changes of expression levels relative to wild-type siblings are indicated (mean ± s.e.m.; n = 3 biological replicas). |
|
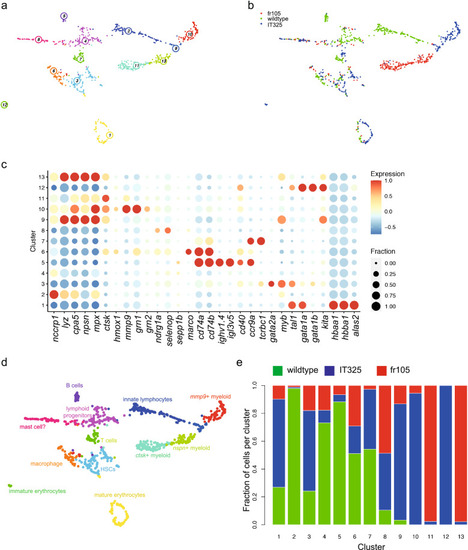
Transcriptional landscape of whole kidney marrow cells of IT325 and fr105 mutants. (a) Uniform manifold approximation and projection (UMAP) representation of transcriptome similarities determined by combined analysis of 341 wild-type, 331 mutant cells of the IT325 line, and 262 mutant cells of the fr105 line; the individual clusters are indicated by colour and numbers. Fish were 3 months of age. (b) Distribution of wild-type and mutant cells among transcriptionally defined cell clusters; origins of cells are colour-coded. (c) Expression pattern of signature genes (listed at the bottom) in the individual cell clusters shown in (a, b). Colour represents the z-score of the mean expression of the gene in the respective cluster and dot size represents the fraction of cells in the cluster expressing the gene. z-scores above 1 and below -1 are replaced by 1 and −1, respectively. (d) Cell type identification based on signature gene expression patterns (see Table 2, and text for details). (e) Distribution of cells of different genotypes in the identified cell clusters. |
|
Transcriptional landscape of whole kidney marrow cells of IT325 mutants. (a) Uniform manifold approximation and projection (UMAP) representation of transcriptome similarities determined by combined analysis of 341 wild-type and 331 mutant cells of the IT325 line; the individual clusters are indicated by colour and numbers. (b) Distribution of wild-type and mutant cells among transcriptionally defined cell clusters; genotypes of cells are colour-coded. (c) Expression pattern of signature genes (listed at the bottom) in the individual cell clusters shown in (a, b). Colour represents the z-score of the mean expression of the gene in the respective cluster and dot size represents the fraction of cells in the cluster expressing the gene. z-scores above 1 and below -1 are replaced by 1 and −1, respectively. (d) Cell type identification based on signature gene expression patterns (see Table 2, and text for details). (e) Graph representation of neighbourhoods identified by the Milo algorithm; the positions of index cells of neighbourhoods are projected onto their positions of the UMAP representation shown in (a, b). Nodes are equivalent to neighbourhoods; colours indicate their compositional differences between wildtype and mutant cells and are quantified as log2 fold changes at FDR 10% (colour code is indicated to the right); the sizes of the nodes denote the number of cells in the neighbourhood; graph edges represent the number of cells shared among adjacent neighbourhoods. (f) The distribution of log2 fold changes in abundance between wildtype and mutant cells in neighbourhoods identified in different cell clusters (identified at the left) is shown in a bee-swarm plot; neighbourhoods exhibiting differential abundance at FDR 10% are colour-coded (reddish colours denote greater abundance of mutant cells; bluish colours denote greater abundance of wildtype cells). |
|
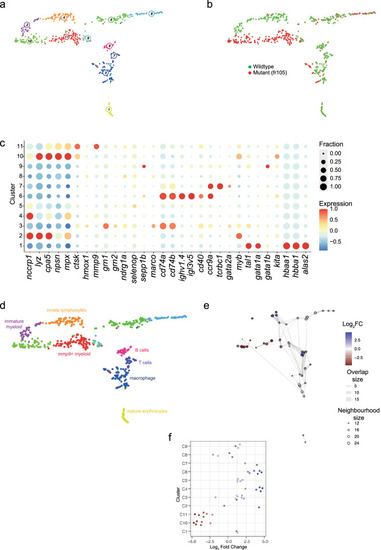
Transcriptional landscape of whole kidney marrow cells of fr105 mutants. (a) Uniform manifold approximation and projection (UMAP) representation of transcriptome similarities determined by combined analysis of 341 wild-type and 262 mutant cells of the fr105 line; the individual clusters are indicated by colour and numbers. (b) Distribution of wild-type and mutant cells among transcriptionally defined cell clusters; origins of cells are colour-coded. (c) Expression pattern of signature genes (listed at the bottom) in the individual cell clusters shown in (a, b). Colour represents the z-score of the mean expression of the gene in the respective cluster and dot size represents the fraction of cells in the cluster expressing the gene. z-scores above 1 and below -1 are replaced by 1 and −1, respectively. (d) Cell type identification based on signature gene expression patterns (see Table 2, and text for details). (e) Graph representation of neighbourhoods identified by the Milo algorithm; see Fig. 3e legend for explanation. (f) Bee-swarm plot of differential abundance analysis; see Fig. 3f legend for further explanation. |