Figure 4
- ID
- ZDB-FIG-221214-22
- Publication
- Hess et al., 2022 - Stage-specific and cell type-specific requirements of ikzf1 during haematopoietic differentiation in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
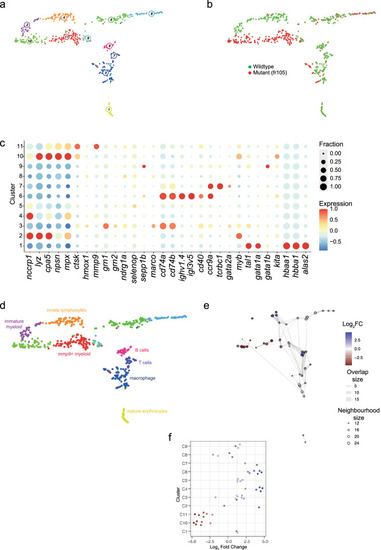
Transcriptional landscape of whole kidney marrow cells of fr105 mutants. (a) Uniform manifold approximation and projection (UMAP) representation of transcriptome similarities determined by combined analysis of 341 wild-type and 262 mutant cells of the fr105 line; the individual clusters are indicated by colour and numbers. (b) Distribution of wild-type and mutant cells among transcriptionally defined cell clusters; origins of cells are colour-coded. (c) Expression pattern of signature genes (listed at the bottom) in the individual cell clusters shown in (a, b). Colour represents the z-score of the mean expression of the gene in the respective cluster and dot size represents the fraction of cells in the cluster expressing the gene. z-scores above 1 and below -1 are replaced by 1 and −1, respectively. (d) Cell type identification based on signature gene expression patterns (see Table 2, and text for details). (e) Graph representation of neighbourhoods identified by the Milo algorithm; see Fig. 3e legend for explanation. (f) Bee-swarm plot of differential abundance analysis; see Fig. 3f legend for further explanation. |