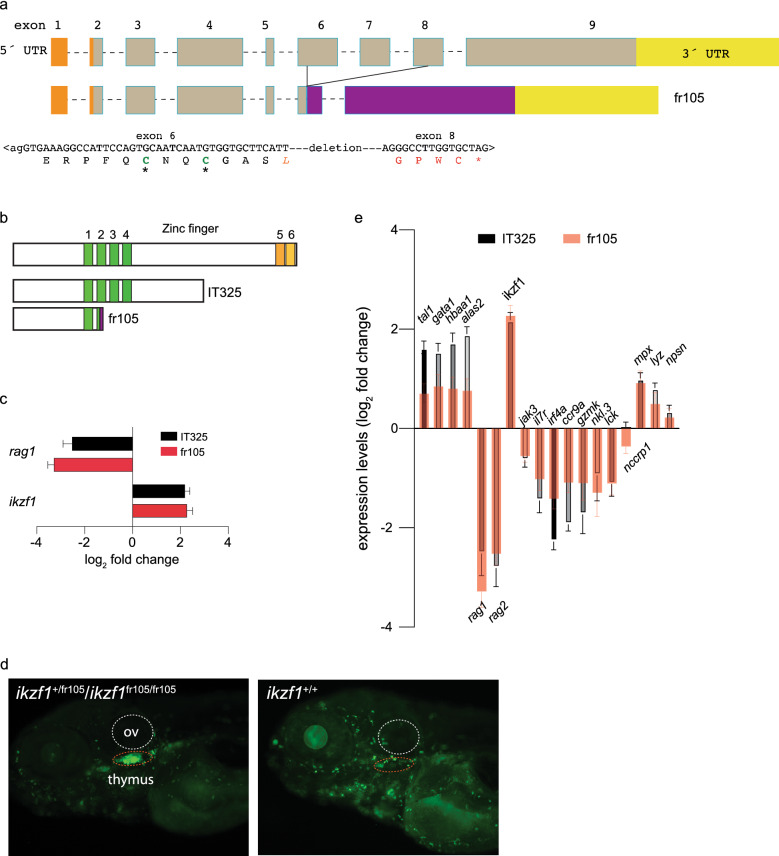
Figure 1 Characterization of a new ikzf1 allele. (a) Schematic structure of the zebrafish ikzf1 gene. Exons are represented by boxes and numbered and drawn to scale; the introns are indicated by dashed lines but are not drawn to scale. The 5´-UTR and 3´-UTR are indicated in orange and yellow. The CRISPR/Cas9-induced deletion in the fr105 allele is indicated; the nucleotide sequence across the junction is shown and the resulting protein sequence is given in single letter code. The deletion creates a frame-shift, resulting in the addition of unrelated amino acid residues (red letters) before a premature stop codon (*). (b) Schematic of the proteins encoded by the two ikzf1 alleles studied here; the IT325 allele was described by Schorpp et al.9. (c) Lack of larval T cell development in homozygous IT325 and fr105 mutants as determined by RNA-seq at 5 dpf; rag1 and ikzf1 expression levels relative to wildtype are shown (mean ± s.e.m.; n = 3 biological replicas). (d) Analysis of the ikzf1fr105 allele. In the ikaros:eGFP background, wild-type and heterozygous fish cannot be phenotypically distinguished (left panel), whereas the mutants (right panel) lack fluorescent signals in the thymus (dashed red oval), and exhibit brighter fluorescence. ov, otic vesicle. e Differential gene expression patterns in IT325 and fr105 mutants at 5 dpf. The log2 fold changes of expression levels relative to wild-type siblings are indicated (mean ± s.e.m.; n = 3 biological replicas).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Sci. Rep.