- Title
-
Putative COVID-19 therapies imatinib, lopinavir, ritonavir, and ivermectin cause hair cell damage: A targeted screen in the zebrafish lateral line
- Authors
- Coffin, A.B., Dale, E., Doppenberg, E., Fearington, F., Hayward, T., Hill, J., Molano, O.
- Source
- Full text @ Front. Cell. Neurosci.
|
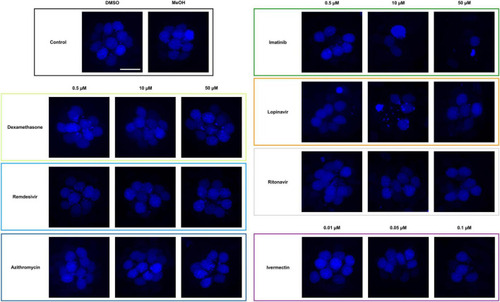
Some COVID-19 drugs are toxic to zebrafish hair cells. Hair cells were live-labeled with DAPI (blue), then fish were treated for 24 h in the indicated drug. The top left panel shows the vehicle controls with a full complement of hair cells. Methanol (MeOH) served as the vehicle for dexamethasone, while DMSO is the vehicle for all other drugs shown here. All other panels show representative confocal images for three concentrations of each drug. Left panels: non-ototoxic drugs dexamethasone, remdesivir, and azithromycin. Right panels: imatinib, lopinavir, ritonavir, and ivermectin all caused hair cell loss. Note that ivermectin was toxic to the animal at high concentrations, so only the non-lethal concentrations were used for this experiment. The scale bar in the upper left = 10 μm and applies to all panels. |
|
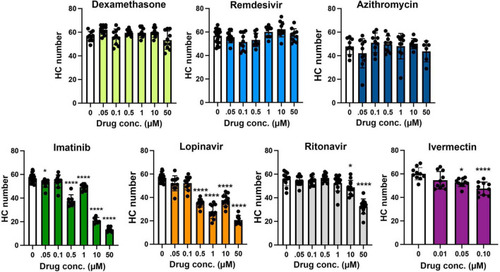
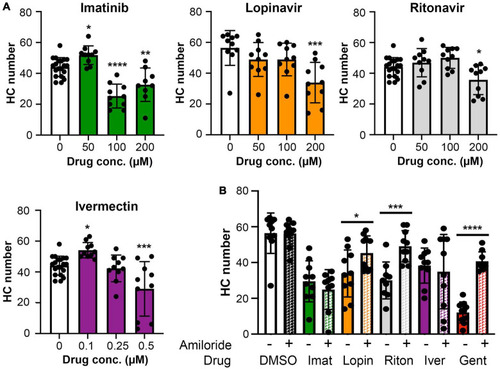
Hair cell quantification following treatment with COVID-19 therapies. Hair cells were counted from images represented in |
|
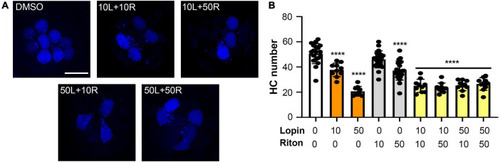
Hair cell toxicity of lopinavir and ritonavir combinations. DAPI-labeled fish were treated for 24 h with 10 and/or 50 μM of lopinavir (L), ritonavir (R), or both drugs. |
|
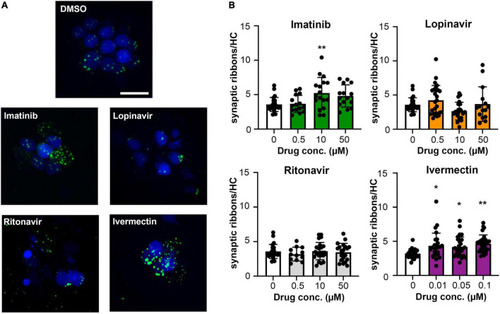
Some COVID-19 drugs alter the number of pre-synaptic ribbons. Rib-GFP fish were live-labeled with DAPI and treated with imatinib, lopinavir, ritonavir, or ivermectin for 24 h. |
|
Most COVID-19 therapies do not cause overall cytotoxicity. |
|
|
|
Ivermectin is not ototoxic to rats |