- Title
-
Tal2 is required for generation of GABAergic neurons in the zebrafish midbrain
- Authors
- Wang, R., Guo, S., Yang, L.
- Source
- Full text @ Dev. Dyn.
|
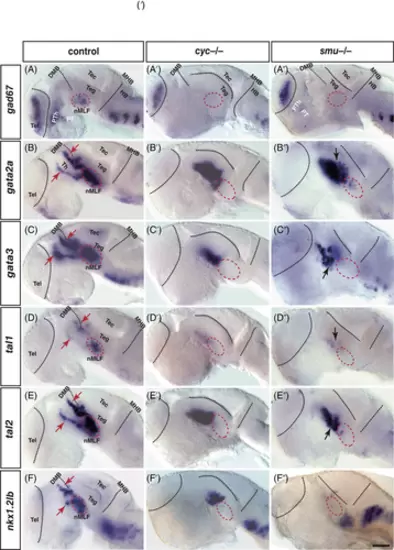
Nodal-related factors and shh signaling are required for gad67, gata2a, gata3, tal1, tal2, and nkx1.2lb expressing in the diencephalon and ventral midbrain. (A-F″) Heads of wild-type (A-F) and matching homozygous cyclops mutants (A′-F′) and smu mutants (A″-F″) of 30 hpf zebrafish embryos hybridized to gad67 (A-A″), gata2a (B-B″), gata3 (C-C″), tal1 (D-D″), tal2 (E-E″), and nkx1.2lb (F-F″) probes. The expression of gad67 (A′) in the diencephalon, and the nucleus of the medial longitudinal fasciculus (nMLF) is abolished in the cyc−/− mutant, while smu−/− embryo (A″) lacks gad67 expressing cells in the nMLF (within the circle). The cyc−/− and smu−/− embryos lack the expressions of gata2a, gata3, tal1, tal2, and nkx1.2lb in the presumptive nMLF, and disturb their expression domains in the diencephalon. All embryos' eyes were removed by dissection. Dorsal is up; rostral, to the left. DMB, diencephalon midbrain boundary; HB, hindbrain; MHB, midbrain hindbrain boundary; nMLF, nucleus of the medial longitudinal fasciculus; PT, posterior tuberculum; PTh, prethalamus; Tec, tectum; Teg, tegmentum; Tel, telencephalon; Th, thalamus. Scale bar: 50 μm |
|
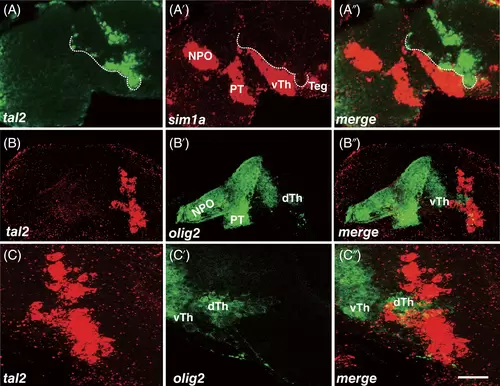
Co-localized analysis of tal2 expressing cells and marker genes in the diencephalon. (A-A″) Heads of wild-type of 30 hpf zebrafish hybridized to tal2 (A) and sim1a (A′) antisense probes and a merged view (A″). The anterior strip of tal2-positive cells is along the boundary of sim1a expressing cells (dotted line). (B-C″) tal2 in situ hybridization (B and C), GFP immunohistochemistry (B′ and C′) and merged views (B″ and C″). Almost all GFP marked olig2 expressing cells are tal2 negative. Of note, the GFP-positive cells located dorsal thalamus extend and separate dorsal and ventral groups of tal2 expressing cells. (C-C″) The high magnification of images. (A-C″) projections of several sections. All embryos' eyes were removed by dissection. Dorsal is up; rostral, to the left. dTh, dorsal thalamus; NPO, neurosecretory preoptic area; PT, posterior tuberculum; Teg, tegmentum; vTh, ventral thalamus. Scale bar: 25 μm in A-B″ and 50 μm in C-C″ |
|
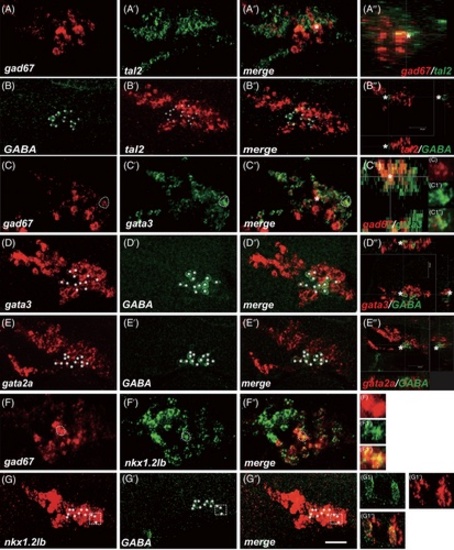
GABAergic neurons co-express tal2, gata3, gata2a, and nkx1.2lb factors in the diencephalon and nMLF. (A-A‴) Heads of wild-type of 30 hpf zebrafish hybridized to gad67 (A) and tal2 (A′) antisense probes, a merged view (A″), and a transverse section at the level of cell marked with asterisk (A‴). gad67 expressing cells are tal2-positive in the diencephalon and nMLF with a varying level. (B-B‴) GABA immunohistochemistry (B), tal2 in situ hybridization (B′), a merged view (B″), and a transverse section at the level of cell marked with asterisk (B‴). Almost all GABAergic neurons co-express tal2 mRNA in the nMLF (plus). (C-C‴) gad67 (C), gata3 (C′) expressing cells, merge (C″), and a transverse section at the level of cell marked with asterisk (C‴). gad67 mRNA expressing cells co-express gata3 at the single cell level (C1 to C1″). (D-D‴) gata3 in situ hybridization (D), GABA immunohistochemistry (D′), a merged view (D″), and a transverse section at the level of cell marked with asterisk (D‴). All GABAergic neurons co-express gata3 mRNA in the nMLF (asterisk). (E-E‴) gata2a in situ hybridization (E), GABA immunohistochemistry (E′), a merged view (E″), and a transverse section at the level of cell marked with asterisk (E‴). Many GABAergic neurons co-express gata2a mRNA in the nMLF (stars). (F-F″) gad67 (F), nkx1.2lb (F′) expressing cells, merge (F″), and views at the level of single cell (F1 to F1″). gad67 mRNA expressing cells co-express nkx1.2lb. (G-G″) nkx1.2lb (G), GABA immunohistochemistry (G′), a merged view (G″), and views at the level of single cell (G1 to G1″). All GABAergic neurons co-express nkx1.2lb mRNA (asterisk). (A-F″) projections of single section. Dashed regions in main panels (C-C′) and (F-G″) are shown in magnified view in panels (C1-G1″). All embryos' eyes were removed by dissection. Dorsal is up; rostral, to the left. Scale bar: 25 μm |
|
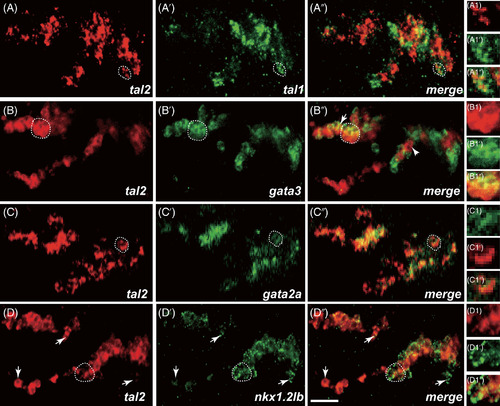
Co-localized analysis of tal2 expressing cells and marker genes in the diencephalon and nMLF. (A-A″) Heads of wild-type of 30 hpf zebrafish hybridized to tal2 (A) and tal1 (A′) antisense probes, a merged view (A″), and views at the single cell level (A1 to A1″). tal2 and tal1 show an overlapping expression domain in the diencephalon and nMLF. (B-B″) tal2 expressing cells (B), gata3 mRNA expression (B′), a merged views (B″), and views at the single cell level (B1 to B1″). Almost all tal2 expressing cells co-express gata3, particularly in the dorsal stripe and nMLF. (C-C″) tal2 mRNA expression (C), gata2a-expressing cells (C′), merge (C″), and views at the single cell level (C1 to C1″). tal2 mRNA expression is identical to gata2a mRNA expression. (D-D″) tal2 (D), nkx1.2lb (D′) mRNA expression domains, merge (D″), and views at the single cell level (D1 to D1″). tal2 expressing cells and nkx1.2lb expressing cells are overlapping in the diencephalon and nMLF. (A-D″) single confocal section. Dashed regions in main panels (A-D″) are shown in magnified view in panels (A1-D1″). Dorsal is up; rostral, to the left. Scale bar: 25 μm |
|
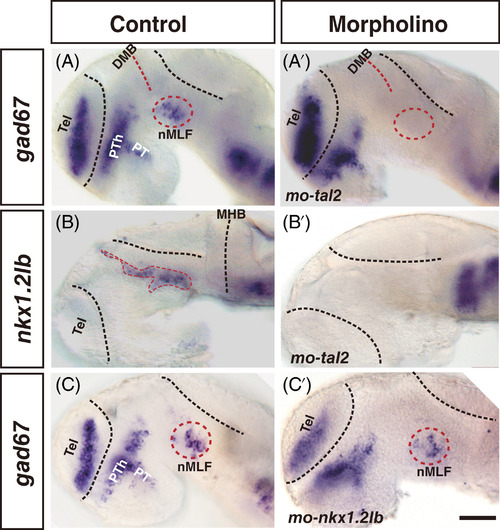
gad67 and nkx1.2lb expression required transcription factor Tal2. (A-B′) Embryos injected with a mismatch control morpholino (A and B) or a morpholino directly targeted tal2 mRNA (A′ and B′). gad67 expressing cells are abolished in the nMLF in morphant (cycle with dash line), whereas gad67-positive cells in the forebrain are present (A′). tal2 knockdown completely eliminates nkx1.2lb expressing cells in the diencephalon and nMLF/nPC, whereas nkx1.2lb expressing cells are unaffected in the hindbrain (B′). (C and C′) Embryos injected with a mismatch control morpholino (C) or a splice morpholino directly targeted exon-intron boundary of nkx1.2lb (C′). The expression of gad67 is unaffected in the nMLF. All embryos' eyes were removed by dissection. Dorsal is up; rostral, to the left. DMB, diencephalon midbrain boundary; nMLF, nucleus of the medial longitudinal fasciculus; PT, posterior tuberculum; Tel, telencephalon; PTh, prethalamus. Scale bar: 50 μm |
|
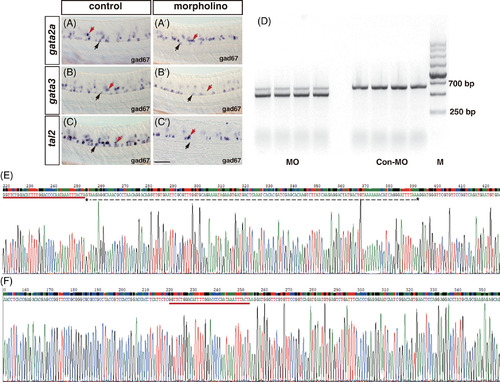
Morpholinos used in this study have high efficiency and specificity. (A-C′) Embryos injected with a mismatch control morpholino (A-C) or a morpholino directly targeted gata2a, gata3, and tal2 mRNA (A′, B′, C′). Injection of gata2a and tal2 morpholinos abolishes gad67 expressing in the KA″ cells. The expression of gad67 in the KA′ cells is eliminated by knockdown of gata3. (D) Injection of splice morpholino directly targeted exon-intron boundary of nkx1.2lb generates a smaller RT-PCR DNA fragment than that of the control. (E and F) DNA sequencing reveals a generation of 137 bp deletion in embryos injected splice morpholino against nkx1.2lb (E, marked by dotted line), in comparison with control (F). Dorsal is up; rostral, to the left. Scale bar: 50 μm |
|
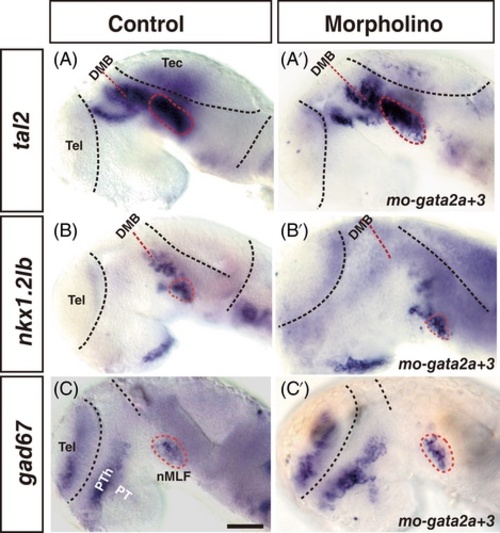
gata2a and gata3 are dispensable for tal2, nkx1.2lb, and gad67 expression. (A-C′) Embryos injected with a mismatch control morpholino (A-C) or morpholinos directly targeted gata2a and gata3 (A′, B′, C′). tal2 (A and A′), nkx1.2lb (B and B′), and gad67 (C and C′) expressing cells are present in the nMLF in the morphants (within cycle). All embryos' eyes were removed by dissection. Dorsal is up; rostral, to the left. DMB, diencephalon midbrain boundary; nMLF, nucleus of the medial longitudinal fasciculus; PT, posterior tuberculum; Tel, telencephalon; PTh, prethalamus. Scale bar: 50 μm |