- Title
-
Congenital Asplenia Interrupts Immune Homeostasis and Leads to Excessive Systemic Inflammation in Zebrafish
- Authors
- Xie, L., Chen, Z., Guo, H., Tao, Y., Miao, X., Wu, R., Li, Y.
- Source
- Full text @ Front Cell Infect Microbiol
|
Effects of congenital asplenia on inflammatory response in zebrafish larvae. |
|
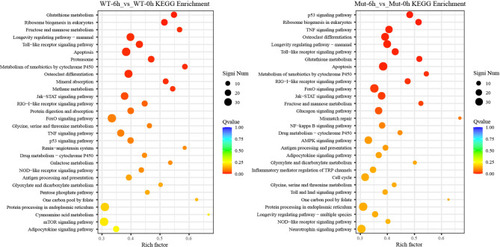
The top 30 enriched KEGG pathways in WT-6h_vs_WT-0h and Mut-6h_vs_Mut-0h. The color and size of the dots indicate Q-values and DEG numbers in pathways, respectively. |
|
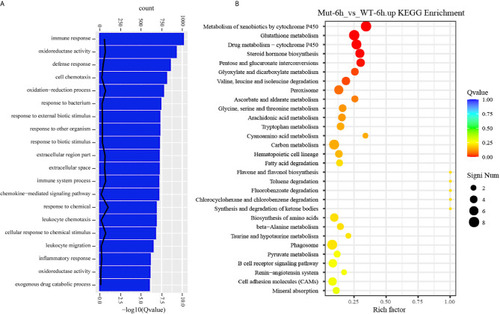
GO functional enrichment and KEGG analysis from upregulated DEGs in Mut-6h_vs_WT-6h. |
|
Congenital asplenia induces excessive systemic inflammation after vaccination in zebrafish. |
|
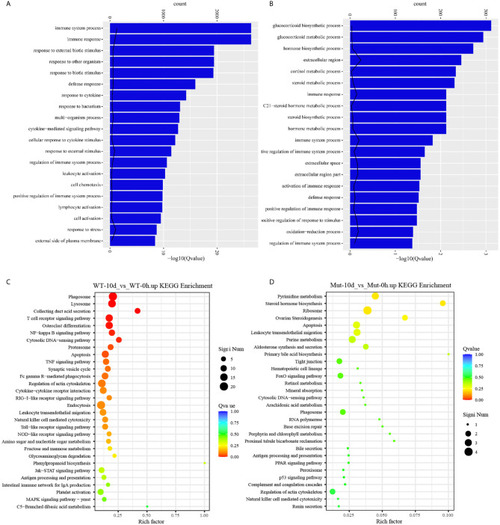
GO functional enrichment and KEGG analysis from upregulated DEGs in WT-10d_vs_WT-0h and Mut-10d_vs_Mut-0h. |
|
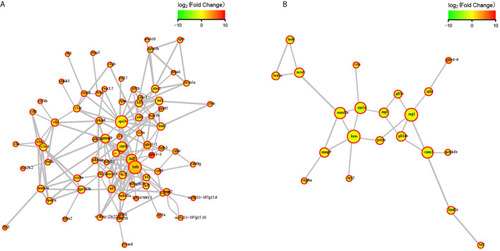
STRING network analysis on DEGs annotated in GO: immune system process. |
|
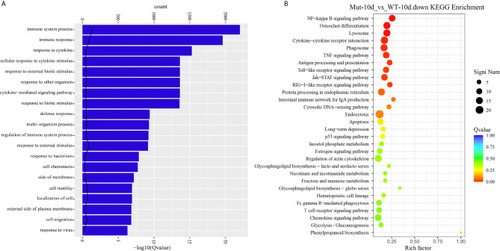
GO functional enrichment and KEGG analysis from downregulated DEGs in Mut-10d_vs_WT-10d. |
|
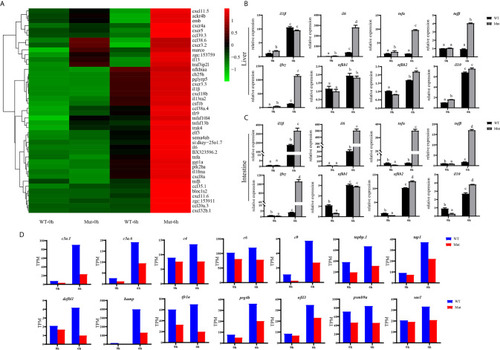
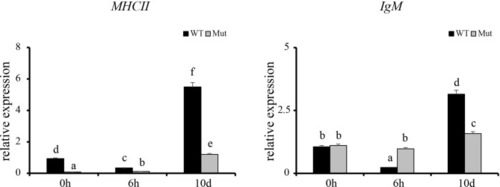
Validation of the 12 DEGs by qRT-PCR. WT animals and congenitally asplenic zebrafish sampled at 0 h were used as controls. The data were normalized to the expression level of WT-0h. The data are reported as the mean ± SEM. Different letters (a, b , c etc.) indicate significant differences ( |
|
Expression profiles of |