- Title
-
RAB13 mRNA compartmentalisation spatially orients tissue morphogenesis
- Authors
- Costa, G., Bradbury, J.J., Tarannum, N., Herbert, S.P.
- Source
- Full text @ EMBO J.
|
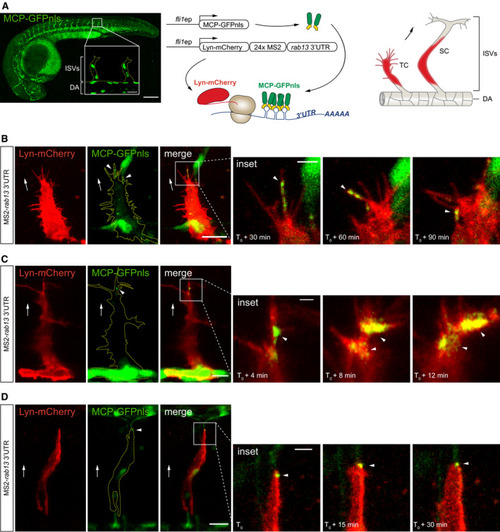
Left: Time‐lapse microscopy of |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|
|
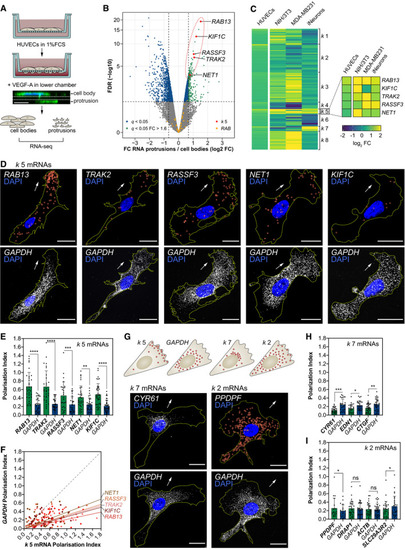
Strategy used to screen for mRNAs enriched in motile protrusions of HUVECs migrating through Transwell membranes. RNAseq data are plotted in log2 fold change (FC) levels of protrusions over cell bodies against adjusted −log10 false discovery rate (FDR) ( Heat map represents the smFISH co‐detection of Polarisation Index (PI) of Top: distribution pattern of mRNAs clustered in PIs of PIs of |
|
Diagram of RNA motif over‐represented in Frequency of mRNAs within each Scheme depicts the Left: representative subconfluent motile cells co‐transfected with plasmids expressing Lyn‐mCherry, MCP‐GFPnls and 24xMS2‐ Left: representative cells co‐transfected with plasmids expressing Lyn‐mCherry, MCP‐GFPnls and 24xMS2‐ |
|
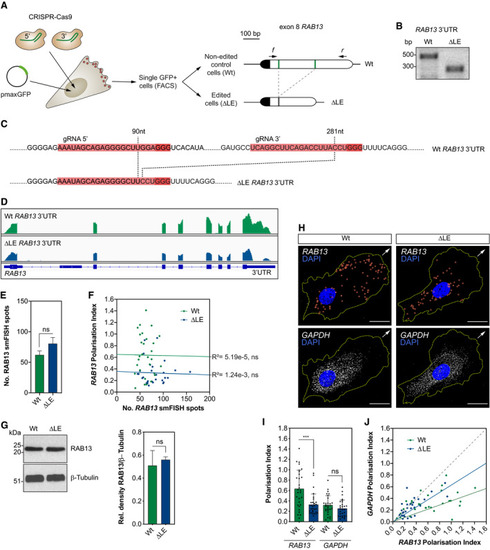
CRISPR‐Cas9 strategy to derive HUVECs with an excision of the LE in the Representative genotyping PCR demonstrates the band size shift in ∆LE HUVECs. Detailed DNA sequence depicting nucleotide positions within the Wt and ∆LE HUVEC RNAseq mapped reads depicting Quantification of Number of Left: representative Western blotting (WB) of Wt and ∆LE HUVECs. Right: densitometry analysis of WB data ( smFISH co‐detection of PI of |
|
Representative time‐lapse microscopy of a bEnd.3 cell co‐transfected with plasmids expressing Lyn‐mCherry, MCP‐GFPnls and 24xMS2‐ Frequency of newly formed filopodia formed within 5‐μm intervals relative to the nearest MCP‐GFPnls particle or a randomised (ctrl) position ( Distance of newly formed filopodia to MCP‐GFPnls or a ctrl position plotted against filopodia duration ( Wt and ∆LE HUVECs co‐cultured on fibroblast monolayers. Endothelial cells were identified either with an antibody against the endothelial cell marker PECAM‐1 (left) or through expression of a nucleofected plasmid encoding the cytoskeletal marker Lifeact‐GFP (right). Number of filopodia detected in co‐cultured HUVECs ( Number of filopodia detected in co‐cultured HUVECs within 12‐μm intervals relative to cell distal tip ( Number of filopodia detected in individual clones of co‐cultured HUVECs within 12‐μm intervals relative to cell distal tip. Illustration of the spatial relationship between |
|
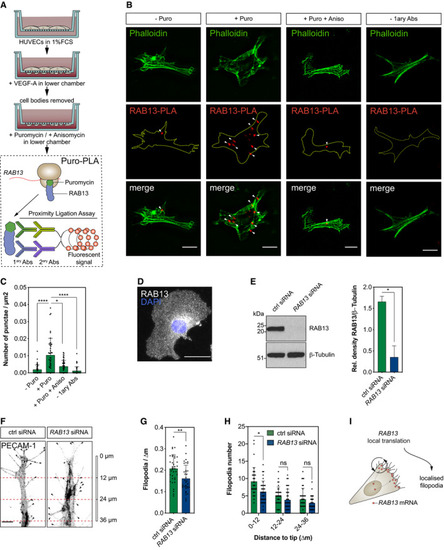
Strategy used to detect local protein synthesis in protrusions formed by HUVECs migrating through Transwell membranes. Representative Puro‐PLA experiments detecting newly synthesised RAB13 in HUVEC protrusions present in the lower side of Transwell membranes. Puro: puromycin; Aniso: anisomycin; 1ary Abs: primary antibodies. Quantification of RAB13 Puro‐PLA punctae normalised to protrusion area ( Representative RAB13 IF assay on migrating HUVECs. Left: representative Western blotting (WB) of siRNA‐transfected HUVECs. Right: densitometry analysis of WB data ( Control (ctrl) and Number of filopodia detected in co‐cultured HUVECs ( Number of filopodia detected in co‐cultured HUVECs within 12‐μm intervals relative to cell distal tip ( Illustration of the spatial relationship between the sites of |
|
CRISPR‐Cas9 strategy to generate the Representative genotyping PCR demonstrates the band size shift in zebrafish harbouring a ∆482 Detailed DNA sequence depicting nucleotide positions within the Wt and ∆482 RNAseq mapped reads depicting qPCR analysis of smFISH detection of Polarisation Index (PI) of EXPRESSION / LABELING:
|
|
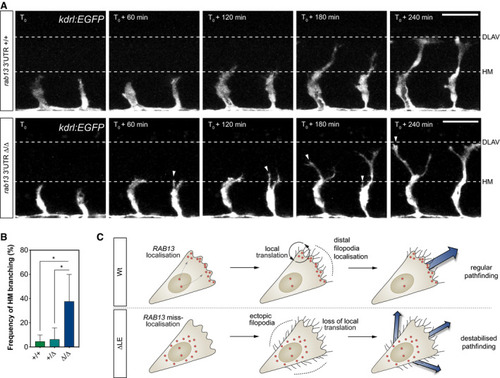
Time‐lapse confocal microscopy of representative Frequency of ISV ectopic branching occurring at the HM ( Illustration of the role for PHENOTYPE:
|
|
Principal component plot depicting the Detail of the heat map shown in Fig Top: distribution pattern of mRNAs clustered in Top: distribution pattern of mRNAs clustered in |
|
Chromatogram confirming the excision of the LE within List of predicted CRISPR‐Cas9 off‐target genes and RNAseq mismatch detection in CRISPR‐Cas9-derived HUVEC clones ( Chromatogram confirming the CRISPR‐Cas9-mediated excision of 482‐nt within the List of predicted CRISPR‐Cas9 off‐target genes and RNAseq mismatch detection in |
|
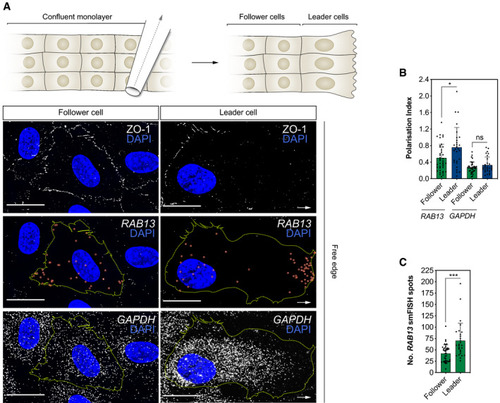
Top: scratch wound assay generates a free edge on a confluent monolayer of HUVECs and encourages cell migration. Bottom: smFISH co‐detection of Polarisation Index of Quantification of the number of |
|
Immunofluorescence analysis of HUVEC protrusions generated on the underside of Transwell membranes and exposed to puromycin after cell body removal. Yellow dashed lines outline protrusion borders; scale bars = 10 μm. |
|
Percentage identity matrix of Multiple sequence alignment between Scheme depicts stages of zebrafish ISV sprouting. DA: dorsal aorta; DLAV: dorsal longitudinal anastomotic vessel; HM: horizontal myoseptum; NC: notochord; NT: neural tube. |