Figure 3
- ID
- ZDB-FIG-201107-17
- Publication
- Costa et al., 2020 - RAB13 mRNA compartmentalisation spatially orients tissue morphogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
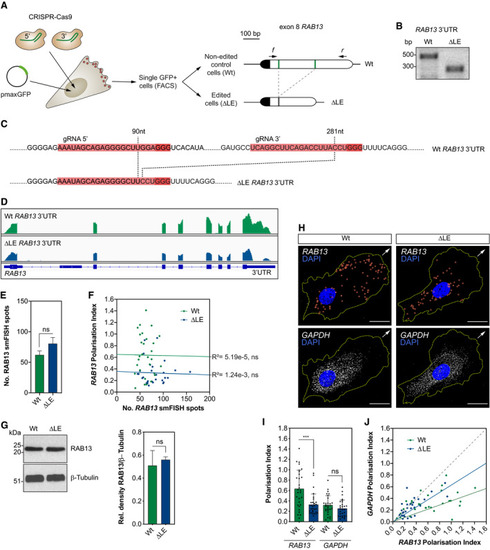
CRISPR‐Cas9 strategy to derive HUVECs with an excision of the LE in the Representative genotyping PCR demonstrates the band size shift in ∆LE HUVECs. Detailed DNA sequence depicting nucleotide positions within the Wt and ∆LE HUVEC RNAseq mapped reads depicting Quantification of Number of Left: representative Western blotting (WB) of Wt and ∆LE HUVECs. Right: densitometry analysis of WB data ( smFISH co‐detection of PI of |