- Title
-
Cardiac sodium channel regulator MOG1 regulates cardiac morphogenesis and rhythm
- Authors
- Zhou, J., Wang, L., Zuo, M., Wang, X., Ahmed, A.S., Chen, Q., Wang, Q.K.
- Source
- Full text @ Sci. Rep.
|
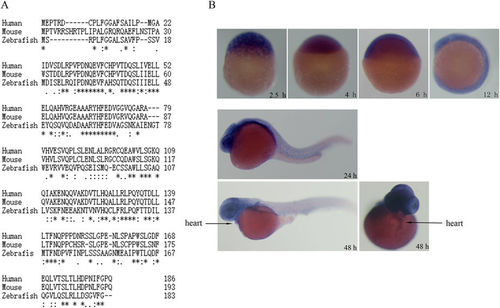
Identification of zebrafish mog1 gene and its expression profile. (A) Alignment of amino acid sequences of human MOG1 (GenBankTM accession number NP_057576), mouse Mog1 (GenBankTM accession number NP_001272370) and zebrafish Mog1 (GenBankTM accession number NP_001093465). (B) Whole-mount in situ hybridization analysis of zebrafish mog1 expression at different developmental stages during embryogenesis. Images for embryos at 2.5 hpf, 4 hpf, 6 hpf and 12 hpf are shown with lateral views with the animal pole to the top. Images for embryos at 24 hpf and 48 hpf stages are lateral views with the head to the left. A dorsal view of the 48 hpf embryo was also shown with the head to the top. |
|
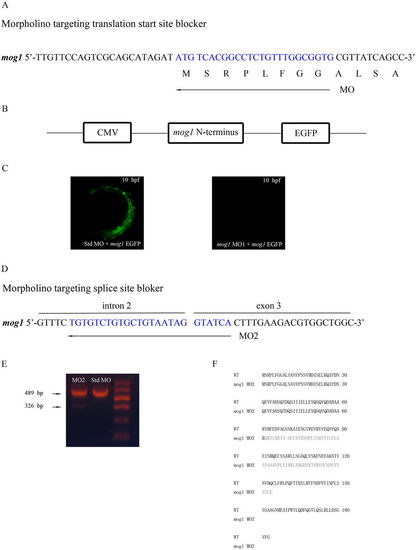
Two mog1 MOs effectively knock expression of mog1 down. (A) Mog1 translation-blocking MO1 was directed against the translation start site. (B) Construction of a pCMV-mog1-EGFP reporter. The 5′-UTR of zebrafish mog1 and a part of the mog1 coding sequence (nucleotide sequences 1–171 starting with A of ATG start codon, amino acid residues 1–57) were fused to the EGFP gene in vector pEGFP-N1, in which expression of the Mog1–EGFP fusion protein (green) is under the control of the CMV promoter. (C) The mog1-EGFP reporter gene was injected into one- to two-cell-stage embryos together with control Std-MO or mog1 MO1. Expression of the Mog1–EGFP fusion protein was examined under a fluorescence microscope at the 10 hpf stage. Note that mog1 MO1 effectively abolished the expression of MOG1–EGFP (lack of green signal) compared with control. (D) Mog1 splicing-blocking MO2 was designed to target the intron 2–exon 3 splicing donor site. (E) Results of RT–PCR analysis with total RNA from 24 hpf embryos treated with MO2 or Std MO. MO2 embryos generated a smaller size of alternatively spliced RT–PCR product (326 bp in MO2 versus 489 bp in Std MO). (F) Sequence analysis of the 326 bp band from MO2-injected embryos revealed that MO2 resulted in an alternatively spliced mog1 transcript with a deletion of exon3 (163 bp). |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
|
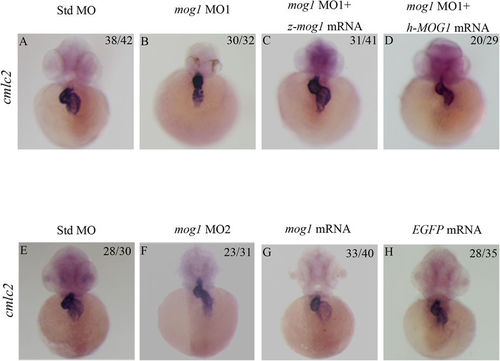
Mog1 regulates cardiac morphogenesis and development during zebrafish embryogenesis. (A,B) Whole-mount in situ hybridization with embryos injected with 16 ng of control Std-MO (A) or 16 ng of mog1 MO1 (B) with a cardiac marker, cmlc2, at 48 hpf. (C,D) Whole-mount in situ hybridization with embryos injected with 16 ng of mog1 MO1 together with 200 pg of zebrafish mog1 mRNA (C) or with 200 pg of human MOG1 mRNA (D) with a cardiac marker, cmlc2, at 48 hpf. Note that overexpression of mog1 rescued the defects in mog1 morphants. The construct for making zebrafish mog1 mRNA was mutated at 5 positions (from ATG TCA CGG CCT CTG TTT to ATG TCT CGT CCG CTA TTC) so that mog1 MO1 can knock endogenous zebrafish mog1 mRNA down, but not the in vitro synthesized mog1 mRNA used for injection. The mog1 MO1 cannot bind to human MOG1 mRNA so that it cannot knock the in vitro synthesized human MOG1 mRNA down. (E,F) Whole-mount in situ hybridization with embryos injected with 16 ng of control Std-MO (E) or 16 ng of mog1 MO2 (F) with a cardiac marker, cmlc2, at 48 hpf. (G,H) Overexpression of mog1 did not cause changes of cardiac phenotype compared with embryos injected with EGFP mRNA (negative controls). The numbers in the upper right corner (e.g. 38/42 in A) describe the ratio of the number of embryos with the phenotype (e.g. 38) in the image over the number of total embryos analyzed (e.g. 42). Scale bar = 100 μM. PHENOTYPE:
|
|
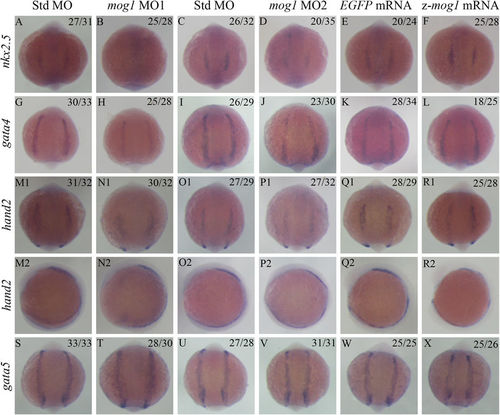
Knockdown of mog1 causes a decrease of expression levels of cardiac transcriptional factors in the anterior lateral mesoderm by whole-mount in situ hybridization. (A–D) Knockdown of mog1 expression reduces intensity of the nkx2.5 signal. Zebrafish embryos (1–2 cell stage) were injected with 16 ng of Std-MO (A,C), 16 ng of mog1 MO1 (B) or 16 ng of mog1 MO2 (D) and used for whole-mount in situ hybridization at 12 hpf with an antisense probe for nkx2.5. (G–J) Knockdown of mog1 expression reduces intensity of the gata4 signal. The embryos were injected and processed as in (A–D), but probed with an antisense probe for gata4. (M–P) Knockdown of mog1 expression reduces expression of hand2. (S–V) Knockdown of mog1 expression does not affect expression of gata5. (E,F,K,L,Q,R,W,X) Over-expression of zebrafish mog1 did not affect expression of cardiac transcriptional factors in the anterior lateral mesoderm at the 12 hpf stage. (A–R1,S–X) Embryos are shown at a dorsal view with the anterior at the top. (M2–R2) embryos are shown in a lateral view with the anterior on the left. The numbers in the upper right corner (e.g. 27/31 in A) describe the ratio of the number of embryos with the phenotype (e.g. 27) in the image over the number of total embryos analyzed (e.g. 31). Scale bar = 100 μM. EXPRESSION / LABELING:
PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cardiac developmental defects in mog1 morphants are rescued by overexpression of nkx2.5. Zebrafish embryos were injected with Std-MO (16 ng; A,E), mog1 MO1 (16 ng; B,F), mog1 MO1 (16 ng) together with in vitro synthesized full-length zebrafish (C) or human MOG1 mRNA (200 pg) (D,G) and used for whole-mount in situ hybridization at 12 somites with antisense probes for nkx2.5 (A–D) or cmlc2 (E–G). Reduced expression of nkx2.5 by knockdown of mog1 was rescued by injection of mog1 mRNA (A–D). Co-injection of nkx2.5 mRNA (G) rescued the cardiac developmental defects in mog1 MO1 morphants (whole-mount in situ hybridization signal for cmlc2). (A–D) Dorsal view with the anterior at the top. (E–G) Anterior view with the ventral side at the top. The numbers in the upper right corner (e.g. 33/37 in A) describe the ratio of the number of embryos with the phenotype (e.g. 33) in the image over the number of total embryos analyzed (e.g. 37). PHENOTYPE:
|