- Title
-
tal1 regulates the formation of intercellular junctions and the maintenance of identity in the endocardium
- Authors
- Schumacher, J.A., Bloomekatz, J., Garavito-Aguilar, Z.V., and Yelon, D.
- Source
- Full text @ Dev. Biol.
|
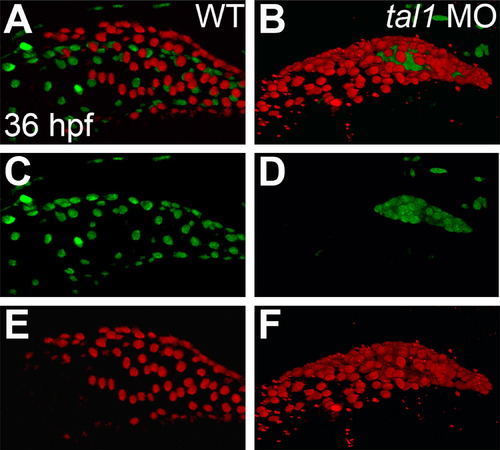
Endocardium fails to populate the atrium in tal1-deficient embryos. (A–F) Lateral views of wild-type (WT) (A, C, and E) and tal1-deficient hearts (B, D, and F) in live embryos at 36 hpf; atrial end of the heart tube to the left. Three-dimensional reconstructions of confocal stacks depict myocardial cells expressing Tg(myl7:H2A-mCherry) (red; A, B, E, and F) and endothelial cells, including the endocardium, expressing Tg(fli1a:negfp) (green; A–D). In wild-type embryos (A and C), the endocardium fully lines the myocardial tube, and extends past the atrium. In tal1-deficient embryos (B and D), the endocardium is clumped within the ventricle (n=151/158). |
|
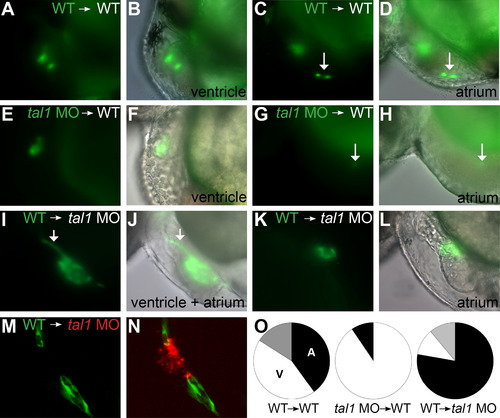
tal1 promotes endocardial extension into the atrium in a cell-autonomous fashion. (A–N) Examples of mosaic embryos generated through blastomere transplantation; lateral views, anterior to the left, at 48 hpf. Overlay of brightfield and fluorescent images indicates location of donor-derived cells. (A–D) Examples of wild-type donor-derived cells expressing Tg(kdrl:GRCFP) (green) in wild-type non-transgenic hosts. (A and B) Wild-type donor-derived cells are located in the ventricular endocardium. (C and D) Wild-type donor-derived cells (arrow) are located in the atrial endocardium. (E–H) Examples of tal1-deficient cells expressing Tg(kdrl:GRCFP) in wild-type non-transgenic hosts. (E and F) tal1-deficient donor-derived cells are located in the ventricular endocardium. (G and H) A single tal1-deficient donor-derived cell (arrow), weakly expressing Tg(kdrl:GRCFP), is located in the atrial endocardium. We found only 2 such examples among our mosaic embryos (Table 1). (I–L) Examples of wild-type cells expressing Tg(kdrl:GRCFP) in tal1-deficient non-transgenic hosts. (I and J) Large number of wild-type donor-derived endocardial cells form a chain of cells through the ventricle (arrow), and extend to line the host atrium. (K and L) Small number of wild-type donor-derived endocardial cells form a small tube of endocardium within the host atrium. (M and N) Example of wild-type cells expressing Tg(kdrl:GRCFP) in a tal1-deficient host expressing Tg(kdrl:HsHRAS-mCherry) (red). Wild-type donor-derived endocardial cells (green) populate the outflow tract and atrium, whereas tal1-deficient host endocardium (red) is clumped within the ventricle. (O) Pie charts indicate the relative contributions of donor-derived cells to the ventricular endocardium (white), atrial endocardium (black), or ventricular and atrial endocardium (gray) within the mosaic embryos that exhibited donor-derived endocardium (Table 1). |
|
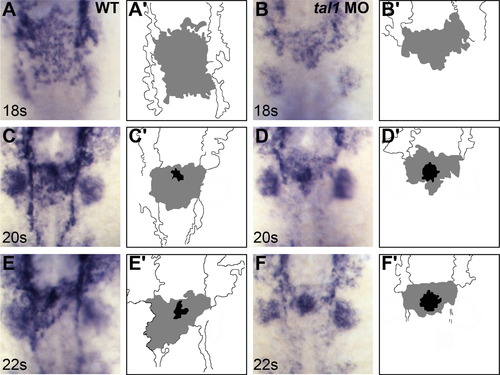
Defects in endocardial sheet formation appear after tal1-deficient endocardial cells arrive at the midline. (A–F) Dorsal views of wild-type (A, C, and E) and tal1-deficient embryos (B, D, and F), anterior to the top, displaying the expression of fli1a at the 18 somite (18s) (A and B), 20s (C and D), and 22s (E and F) stages. (A′–F′) Cartoons indicate regions of fli1a expression in the endocardium (gray and black) and cranial endothelium (white). fli1a expression in the branchial arches is visible in the images (A–F) but is not indicated in the cartoons (A′–F′). (A, C, and E) In wild-type embryos, an endocardial sheet spreads along the anterior-posterior axis by 18s (A). Increased expression of fli1a becomes apparent at the future apex (black areas in C′ and E′) of the endocardial tube as the endocardial sheet transforms into a tube extending toward the left side of the embryo (C and E). (B, D, and F) In tal1-deficient embryos, the expression of fli1a is maintained normally, consistent with previous reports (Patterson et al., 2005). Instead of spreading out into a sheet, the tal1-deficient endocardial cells aggregate at the midline (B) and subsequently fail to migrate toward the left (D and F). EXPRESSION / LABELING:
PHENOTYPE:
|
|
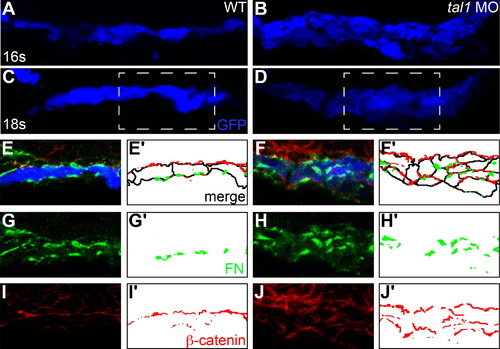
The architecture of the early endocardium is disrupted in tal1-deficient embryos. (A–J) Transverse sections of wild-type (A, C, E, G, and I) and tal1-deficient embryos (B, D, F, H, and J), dorsal to the top, at 16s (A and B) and 18s (C–J). Immunofluorescence detects localization of fibronectin (FN; green) (E–H) and β-catenin (red) (E, F, I, and J); in addition, an anti-GFP antibody allows visualization of Tg(kdrl:GRCFP) expression (blue) in the endocardium (A–F). Together, these markers highlight the shapes and arrangement of the endocardial cells. Endocardial cells form a single-layered sheet of cells at 16s (A), after they have migrated to the midline, and this sheet of cells is maintained at 18s (C). In tal1-deficient embryos, endocardial cells stack on top of each other, forming a multi-layered cluster (B and D). The boxed areas in (C and D) are highlighted in (E–J). The apical-basal polarity of wild-type endocardial cells (E) is indicated by basal deposition of FN (G) and apical enrichment of β-catenin (I). In tal1-deficient embryos (F), localization of FN (H) and β-catenin (J) highlight the dysmorphic endocardial architecture and aberrant cell shapes. (E′–J′) Cartoons indicate regions of FN (green) and β-catenin (red) localization within the endocardium. Endocardial cell outlines are marked in black. The FN and β-catenin that are presumed to be located within the endoderm that overlies the endocardium are excluded from the cartoons. EXPRESSION / LABELING:
PHENOTYPE:
|
|
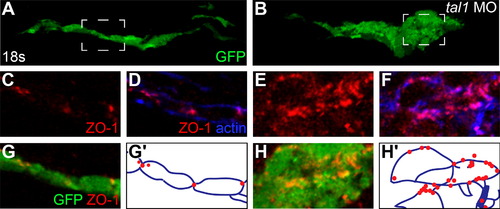
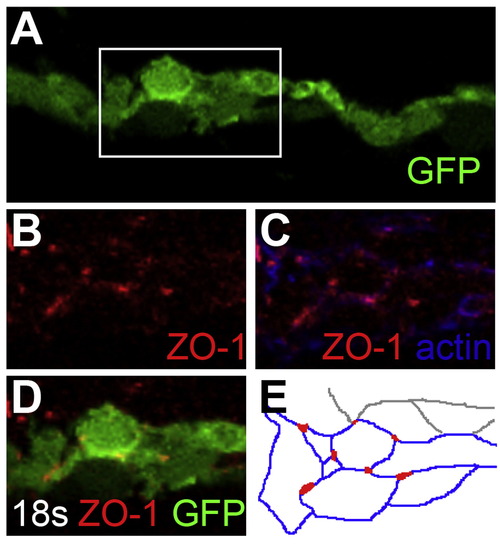
Endocardial intercellular junctions are disorganized following heart tube extension in tal1-deficient embryos. (A–D) Immunofluorescence detects localization of ZO-1 (red) in wild-type (A and C) and tal1-deficient (B and D) hearts at 25 hpf; lateral views, ventricular end of the heart tube at the top. An anti-GFP antibody allows visualization of Tg(kdrl:GRCFP) expression (green) in the endocardium. (A and B) Three-dimensional reconstructions of confocal stacks demonstrate that ZO-1 is located around the perimeter of the myocardial cells in both wild-type and tal1-deficient hearts. (C) Optical slice of a wild-type heart reveals that ZO-1 is localized in discrete puncta at points of endocardial cell-cell contact; in addition, wild-type endocardial cells exhibit a flattened and elongated morphology. (D) Optical slice of a tal1-deficient heart reveals that ZO-1 is mislocalized in large clusters around the perimeter of the endocardial cells; in addition, the tal1-deficient endocardial cells exhibit a rounded shape, rather than the normal elongated shape. (E and F) Immunofluorescence detects ZO-1 (purple) in wild-type (E) and tal1-deficient (F) hearts at 25 hpf; lateral views, ventricular end of the heart tube at the top. Expression of the Tg(fli1a:negfp) (green) and Tg(myl7:H2A-mCherry) (red) transgenes reveals one endocardial cell with ectopic myocardial gene expression (arrow). Subcellular localization of ZO-1 is aberrant in the majority of tal1-deficient endocardial cells, nearly all of which do not display ectopic myocardial gene expression. |
|
Endocardial intercellular junctions are disorganized during early endocardial morphogenesis in tal1-deficient embryos. (A–H) Transverse sections of wild-type (A, C, D, and G) and tal1-deficient embryos (B, E, F, and H), dorsal to the top, at 18s. Immunofluorescence detects localization of ZO-1 (red) (C–H) and F-actin (blue) (D and F); in addition, an anti-GFP antibody allows visualization of Tg(kdrl:GRCFP) expression (green) in the endocardium (A, B, G, and H). The boxed areas in (A and B) are highlighted in (C–H). (A) Wild-type endocardium forms a single-layered sheet with ZO-1 localized to discrete puncta at cell-cell contacts (C, D, and G). (B) tal1-deficient endocardium is multi-layered and disorganized, and ZO-1 is mislocalized in multiple puncta along the endocardial cell boundaries (E, F, and H). (G′ and H′) Cartoons indicate regions of ZO-1 (red) and F-actin (blue) localization within the endocardium. EXPRESSION / LABELING:
PHENOTYPE:
|
|
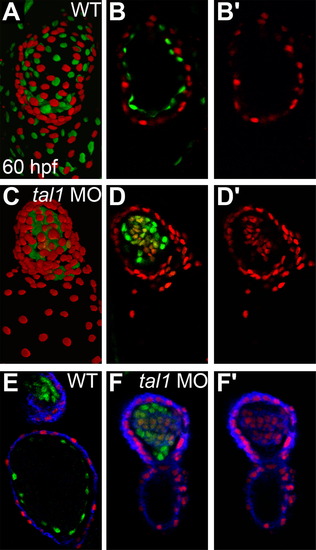
Myocardial genes are ectopically expressed in the tal1-deficient endocardium at 60 hpf. (A–D) Three-dimensional reconstructions of confocal stacks (A and C) and single optical sections through the ventricle (B and D) depict wild-type (A and B) and tal1-deficient hearts (C and D) expressing Tg(myl7:H2A-mCherry) (red) and Tg(fli1a:negfp) (green) at 60 hpf, arterial pole at the top. Many tal1-deficient endocardial cells express both Tg(myl7:H2A-mCherry) and Tg(fli1a:negfp) (D), but wild-type endocardial cells express only Tg(fli1a:negfp) (B). (E and F) Immunofluorescence with the monoclonal antibody MF20 detects sarcomeric myosin heavy chain localization (blue) in wild-type (E) and tal1-deficient hearts (F) expressing Tg(myl7:H2A-mCherry) (red) and Tg(fli1a:negfp) (green). Optical sections through the ventricle reveal ectopic sarcomeric myosin heavy chain within the tal1-deficient endocardium (F). In contrast, sarcomeric myosin heavy chain is not found in the wild-type endocardium (E); section shown depicts both the atrial endocardium (bottom) and outflow tract endocardium (top). (B′, D′, and F′) Images display only the myocardial markers from the sections shown in (B, D, and F). |
|
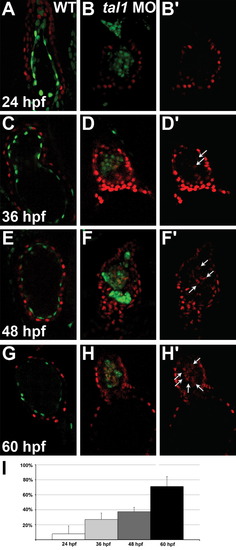
Ectopic myocardial gene expression progressively increases within the tal1-deficient endocardium. (A–H) Optical sections from confocal stacks of wild-type (A, C, E, and G) and tal1-deficient (B, D, F, and H) hearts expressing Tg(myl7:H2A-mCherry) (red) and Tg(fli1a:negfp) (green) at 24 hpf (A and B), 36 hpf (C and D), 48 hpf (E and F) and 60 hpf (G and H), arterial pole at the top. These representative hearts demonstrate the progressive increase in the percentage of endocardial cells expressing Tg(myl7:H2A-mCherry) that occurs between 24 and 60 hpf in tal1-deficient embryos (B, D, F, and H). In contrast, the wild-type endocardium never expresses Tg(myl7:H2A-mCherry) (A, C, E, and G). (B′, D′, F′, and H′) Images display only the myocardial marker from the sections shown in (B, D, F, and H). Arrows indicate examples of endocardial cells ectopically expressing Tg(myl7:H2A-mCherry). (I) Bar graph depicts the average percentage of tal1-deficient endocardial cells expressing Tg(myl7:H2A-mCherry) at each stage examined. Error bars indicate standard deviation (n=4–8 embryos per stage). |
|
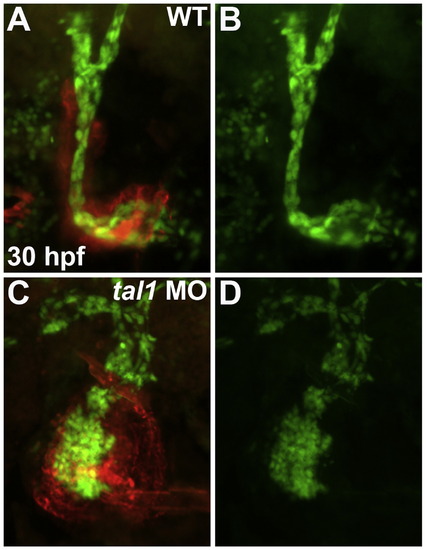
tal1-deficient embryos contain a normal number of endocardial cells. Frontal views of wild-type (A and B) and tal1-deficient hearts (C and D) at 30 hpf; ventricular end of the heart tube at the top. The transgene Tg(fli1a:negfp) (green) was used to count the number of endocardial cells residing within the boundaries of the myocardium, which was labeled by MF20 immunofluorescence (red). |
|
Cell–cell junctions are properly localized in wild-type endocardial cells that are stacked on top of one another. (A–D) Transverse section of wild-type endocardium, dorsal to the top, at 18s. Immunofluorescence detects localization of ZO-1 (red) (B–D) and F-actin (blue) (C); in addition, an anti-GFP antibody allows visualization of Tg(kdrl:GRCFP) expression (green) in the endocardium (A and D). The boxed area in (A) is highlighted in (B–D). Even when wild-type cells are stacked on top of each other, ZO-1 is properly localized to individual vertices at cell-cell junctions (B–D), rather than being found in large patches as in tal1-deficient embryos (Fig. 6E, F, and H). (E) Cartoon indicates regions of ZO-1 (red) and F-actin (blue) localization within the endocardium. |
|
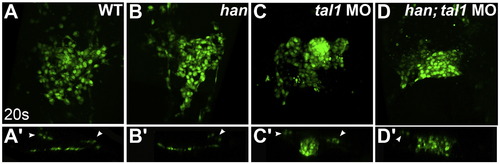
Endocardial aggregation in tal1-deficient embryos does not require hand2 function. (A–D) Three-dimensional reconstructions of confocal stacks depict dorsal views of wild-type (A), han mutant (B), tal1-deficient (C) and han mutant; tal1-deficient (D) embryos expressing Tg(fli1a:negfp) (green) at 20s. (A′–D′) Reconstructions of optical sections displaying transverse slices through the endocardium of the embryos shown in (A–D). Arrowheads mark progenitors of the lateral dorsal aortae. The endocardium forms a single-layered sheet in wild-type (A and A′) and han mutant (B and B′) embryos. In contrast, the endocardium in embryos that are deficient for both han and tal1 (D and D′) forms a multi-layered aggregate similar to that seen in tal1-deficient embryos (C and C′). |
Reprinted from Developmental Biology, 383(2), Schumacher, J.A., Bloomekatz, J., Garavito-Aguilar, Z.V., and Yelon, D., tal1 regulates the formation of intercellular junctions and the maintenance of identity in the endocardium, 214-226, Copyright (2013) with permission from Elsevier. Full text @ Dev. Biol.