- Title
-
Heritable and lineage-specific gene knockdown in zebrafish embryo
- Authors
- Dong, M., Fu, Y.F., Du, T.T., Jing, C.B., Fu, C.T., Chen, Y., Jin, Y., Deng, M., and Liu, T.X.
- Source
- Full text @ PLoS One
|
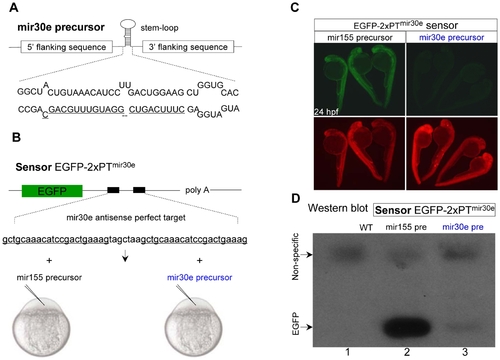
Efficient inhibition of EGFP expression by zebrafish mir-30e precursor in vivo. (A, B) Diagram of zebrafish miR-30e precursor sequence (A), and the EGFP sensor EGFP-2xPTmir30e containing two tandem perfectly complementary target sites for miR-30e binding (2xPT) in its 3′UTR (B, upper). The capped EGFP sensor mRNAs were co-injected with either miR-30e or miR-155 precursor mRNAs into one-cell stage embryos (B, bottom). (C) A dramatic decrease of EGFP fluorescence in 24 hpf embryos co-injected with EGFP-2xPTmir30e sensor (10 pg) and miR-30e precursor (50 pg). The DsRed mRNA was co-injected as an injection control (bottom panels). (D) Western blot analysis of 24 hpf embryos injected with EGFP-2xPTmir30e sensor and miR-155 precursor or miR-30e precursor. The upper non-specific bands were shown as a loading control. |
|
Knockdown of EGFP sensors by mir-shRNA in vivo. (A) Diagram of miRNA-based shRNAEGFP-ORF (mir-shRNAEGFP-ORF). The stem-loop region of miR-30e precursor was replaced with chemically synthesized shRNAEGFP-ORF oligonucleotides containing the same sequence as the miR-30e stem-loop, except that the miR-30e hairpin stem was changed to the sequence that was complementary to the open reading frame (ORF) of EGFP transcript. (B) Northern blot analysis of 12 and 24 hpf embryos injected with in vitro synthesized mir-shRNAEGFP-ORF mRNAs. The U6 promoter-driven expression of shRNAEGFP-ORF in 293T cells was used as a positive control (left lanes). (C) Diagram of various sensors containing one or two copies of binding sequence for mir-shRNAEGFP-ORF within the ORF or the 3′UTR. The 22-bp long binding sequence was inserted into the 3′UTR-SV40 of either EGFP or DsRed gene. (D) Individual capped sensor mRNAs was co-injected with either miR-30e precursor control or mir-shRNAEGFP-ORF. (E–H) Detection of EGFP and DsRed fluorescence in 24 hpf embryos injected with various sensor mRNAs. Red fluorescence was used as an injection control in E, F and G, and green fluorescence as injection control in H. (I) Western blot analysis of 24 hpf embryos shown in panels E–G. The β-actin was used as a loading control. |
|
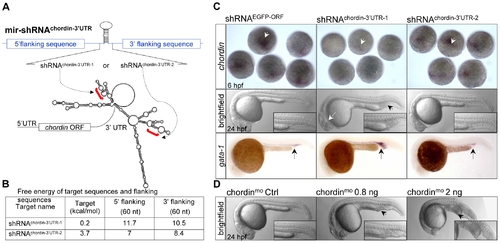
Knockdown of endogenous cellular chordin expression. (A) Diagram of mir- shRNAchordin-3′UTR-1 and mir-shRNAchordin-3′UTR-2 against the 3′UTR of chordin gene, whose predicted secondary structure was shown at the bottom. Red brackets denoted the targeted regions. (B) Free-energy of the targeted regions and corresponding flanking sequence predicted with mFold software. (C) Phenotypes of chordin-deficient embryos. WISH analysis of chordin expression in the 6 hpf embryos injected with 200 pg of shRNAEGFP-ORF (control), shRNAchordin-3′UTR-1 or shRNAchordin-3′UTR-2 mRNAs using a dig-labeled full-length 3′UTR of chordin as probe (upper panels). A significantly enlarged ICM (black arrowhead) with a partial loss of neural tissues (white arrow, 61/99) were observed in 24 hpf embryos injected with only shRNAchordin-3′UTR-1, but not with shRNAchordin-3′UTR-2 and control (middle panels). A higher level of gata-1 expression was also detected in the shRNAchordin-3′UTR-1-injected embryos, compared to control or shRNAchordin-3′UTR-2-injetced embryos (bottom panels). (D) Morphology of embryos injected with chordin morpholino oligonucleotides and its 4-base pair mismatch control. Embryos at 6 hpf are dorsal view, and embryos at 24 hpf are lateral views with head to the left. |
|
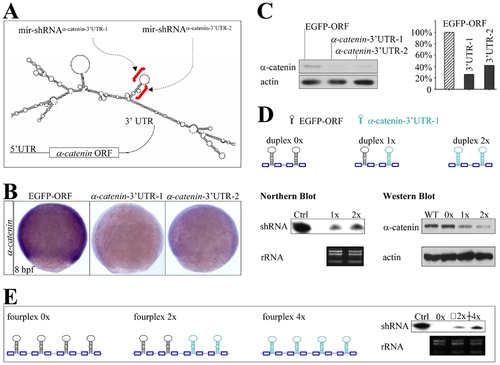
Knockdown of endogenous α-catenin expression. (A) Diagram of mir-shRNAα-catenin-3′UTR-1 and mir-shRNAα-catenin-3′UTR-2 against the 3′UTR of α-catenin gene, whose predicted secondary structure was shown at the bottom. Red brackets denoted the targeted regions. (B) WISH analysis of α-catenin expression in the 8 hpf embryos injected with 160 pg of either shRNAEGFP-ORF, shRNAα-catenin-3′UTR-1 or shRNAα-catenin-3′UTR-2 mRNAs. (C) Quantitative Western blot analysis of α-catenin protein in 22 hpf embryos injected with shRNAs shown in panels B (5 embryos for each lane). (D) Diagram of mir-shRNA duplexes carrying two copies of shRNAEGFP-ORF (duplex 0x), one copy for each shRNAEGFP-ORF and shRNAα-catenin-3′UTR-1 (duplex 1x), and two copies of shRNAα-catenin-3′UTR-1 (duplex 2x). Northern and Western blot analyses of 22 hpf embryos injected with shRNAs 0x, 1x or 2x as shown at the bottom (100 embryos for each line). The ribosomal 5S RNA and β-actin was used as a loading control, respectively. (E) Diagram of mir-shRNA fourplexes carrying four copies of shRNAEGFP-ORF (0x), two copies for each shRNAEGFP-ORF and shRNAα-catenin-3′UTR-1 (2x), and four copies of shRNAα-catenin-3′UTR-1 (4x). Northern blot analysis of 22 hpf embryos injected with shRNAs 0x, 2x or 4x (100 embryos for each line). The ribosomal RNA was used as a loading control. |
|
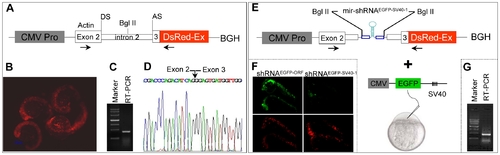
Design of pol II promoter-driven knockdown construct. (A) Diagram of pol II-type promoter CMV driven-knockdown vector (CMV promoter-actin-DsRed-BGH). DS: donor site; AS: acceptor site. The first 21-base pairs of exon 3 of zebrafish actin gene have been in-frame fused to the DsRed fluorescent protein gene followed by a bovine growth hormone (BGH) sequence as 3′UTR. (B) Red fluorescence was observed in 22 hpf embryos injected with the plasmid shown in panel A. (C) RT-PCR analysis with total RNAs derived from 22 hpf embryos shown in panel B. The primers used are indicated by horizontal arrows in panel A. (D) The sequence of RT-PCR product shown in pane C. Note that the entire intron 2 of the actin gene has been spliced out (arrow). (E) The mir-shRNAEGFP-SV40-1 was inserted into the intron 2 at the Bgl II site and co-injection with CMV-EGFP-SV40 reporter plasmid. (F) Knockdown of EGFP fluorescence in the 24 hpf embryos co-injected with EGFP-SV40 reporter plasmid plus CMV promoter-actin-DsRed-BGH plasmid carrying either mir-shRNAEGFP-ORF or mir-shRNAEGFP-SV40-1. (G) RT-PCR analysis of total RNAs derived from 22 hpf embryos shown in panel F. |
|
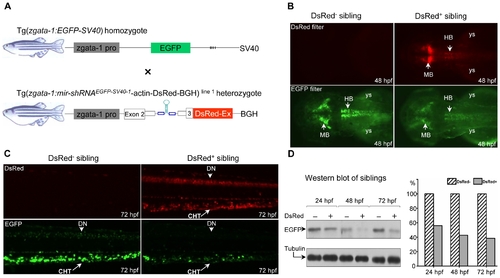
Tissue-specific knockdown of chromosomally integrated EGFP expression. (A) Diagram of transgenic lines Tg(zgata-1:EGFP-SV40) and Tg(zgata-1:mir-shRNAEGFP-SV40-1-actin-DsRed-BGH) line 1 under control of zebrafish gata-1 promoter. (B) Knockdown of EGFP fluorescence was observed in the mid- and hindbrain of the DsRed+, but not DsRed- F2 sibling at 48 hpf. Embryos are dorsal view with head to the left. (C) Knockdown of EGFP fluorescence was observed in the dorsal neurons and caudal hematopoietic tissue of the DsRed+, but not DsRed- F2 sibling at 72 hpf. MB: midbrain; HB: hindbrain; ys: yolk sac; DN: dorsal neurons; CHT: caudal hematopoietic tissue. Embryos are lateral view with head to the left. (D) Western blot analysis of EGFP expression in the DsRed- and DsRed+ F2 embryos at 24, 48 and 72 hpf. The α-tubulin protein was used as a loading control. |
|
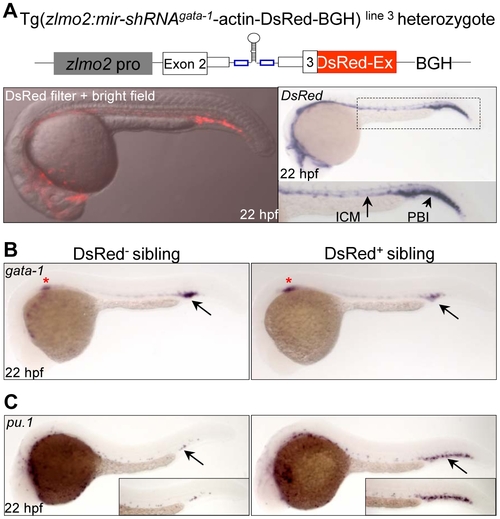
Tissue-specific knockdown of endogenous gata-1 expression. (A) The expression of DsRed fluorescence and transcripts recapitulates the endogenous lmo2 expression pattern in the Tg(zlmo2:mir-shRNAgata-1-actin-DsRed-BGH) line 3. Note that stronger expression of DsRed transcripts can be observed in the PBI region than in the ICM region. ICM: intermediate cell mass; PBI: posterior blood island. (B) WISH analysis of gata-1 expression in the DsRed- and DsRed+ F1 siblings at 22 hpf. Red star denotes the gata-1 staining in the developing kidney. (C) WISH analysis of pu.1 expression in the DsRed- and DsRed+ F1 siblings at 22 hpf. Note that the pu.1 staining is massively increased (arrow). All embryos are dorsal view with head to the left. |
|
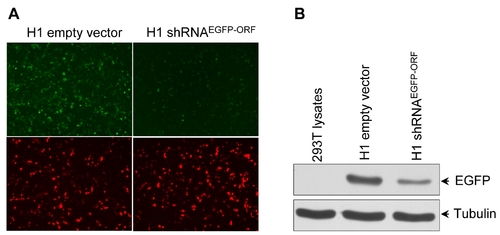
Knockdown of EGFP gene by H1 pol III promoter shRNAEGFP-ORF in cultured 293T cells. (A) EGFP fluorescence and (B) Western blot analysis of 48 hours post transfection. |
|
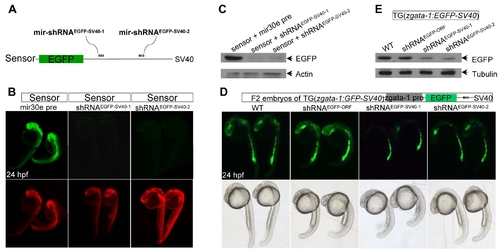
Transient knockdown of chromosomally integrated EGFP gene. (A) Diagram of mir-shRNAEGFP-SV40-1 and mir-shRNAEGFP-SV40-2 against the proximal and distal SV40-3′UTR of EGFP, respectively. (B) Detection of EGFP and DsRed fluorescence in 24 hpf embryos injected with indicted mRNAs. Red fluorescence was used as an injection control. (C) Western blot analysis of 24 hpf embryos shown in panels B. The β-actin was used as a loading control. (D) Detection of EGFP fluorescence in the Tg(zgata-1:EGFP-SV40) transgenic embryos injected with indicated mRNAs. The development and morphology of injected embryos appeared to be normal (bottom panels). (E) Western blot analysis of 24 hpf embryos shown in panels D. The α-tubulin was used as a loading control. |
|
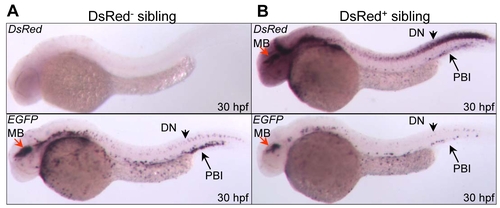
Tissue-specific knockdown of chromosomally integrated EGFP transcripts. WISH analysis of EGFP and DsRed mRNA expression in DsRed- (A), and DsRed+ embryos (B) at 30 hpf. MB: midbrain; DN: dorsal neuron; PBI: posterior blood island. |