- Title
-
Nlz1/Znf703 acts as a repressor of transcription
- Authors
- Nakamura, M., Choe, S.K., Runko, A.P., Gardner, P.D., and Sagerström, C.G.
- Source
- Full text @ BMC Dev. Biol.
|
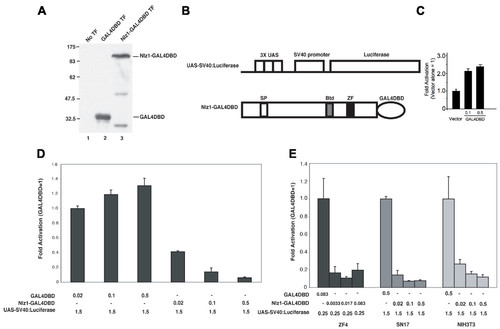
Nlz1 represses transcription in various cell lines. A. Western blot demonstrating that GAL4DBD (lane 2) and the Nlz1-GAL4DBD fusion protein (lane 3) are well expressed. No TF = untransfected control. B. Schematic outline of constructs used. The reporter construct (Top) contains three UAS elements (GAL4 binding sites) upstream of the SV40 promoter and the Luciferase reporter gene. The Nlz1-GAL4DBD construct (bottom) was generated by fusing the GAL4DBD in frame to the C-terminus of full-length zebrafish Nlz1. SP = Sp domain; Btd = buttonhead domain; ZF = C2H2 zinc finger domain. C-E. HeLa cell reporter assays were carried out as described in the Methods section. Luciferase activity was normalized to β-galactosidase activity and the data are presented as fold activation relative to control vector. C. The Gal4DBD domain is a weak activator. D. Nlz1 represses transcription from the UAS-SV40:Luciferase reporter in HeLa cells. E. Nlz1 represses transcription in ZF4, SN17 and NIH3T3 cells. |
|
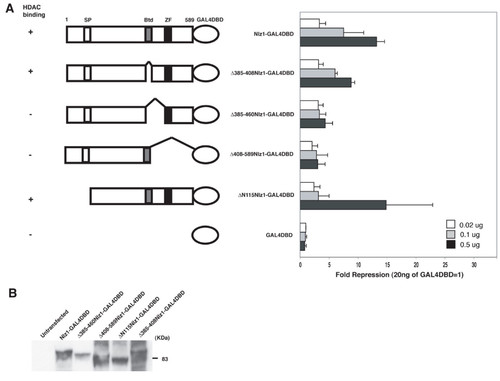
Nlz1-mediated repression requires an intact HDAC interaction domain. A. Schematic outline of deletion constructs used in reporter assay. SP = Sp domain; Btd = Buttonhead domain; ZF = C2H2 zinc finger domain. Column at left indicates binding of each construct to HDACs as reported previously [3]. Graph at right indicates the ability of each construct to repress the UAS-SV40:Luciferase reporter in HeLa cells. Transfection was carried out as in Fig. 1 using DNA doses as indicated. Data are presented as fold repression relative to 20 ng of GAL4DBD alone. B. Western blot demonstrating that each construct is well expressed in HeLa cells. |
|
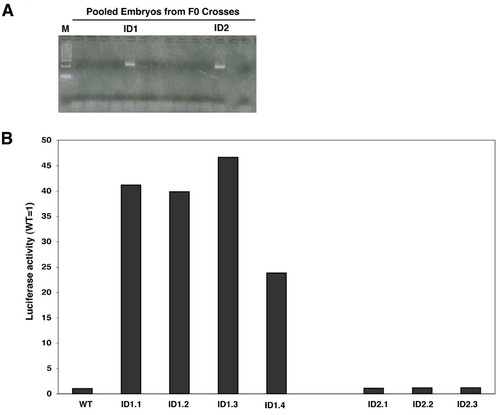
Derivation of the stable transgenic TG(UAS-SV40:Luciferase) zebrafish reporter line.A. Potential F0 founder fish were identified by PCR as outlined in the Methods section. Two founders (ID1 and ID2) were identified. B. Expression of the Luciferase reporter in stable transgenic embryos. TG(UAS-SV40:Luciferase) F1 carriers were out-crossed and their offspring assayed for luciferase activity. Four F1 carriers derived from the ID1 founder (ID1.1, ID1.2, ID1.3 and ID 1.4) produced embryos with robust luciferase activity, while 3 carriers derived from the ID2 founder (ID2.1, ID2.2 and ID2.3) produced embryos lacking luciferase activity. EXPRESSION / LABELING:
|
|
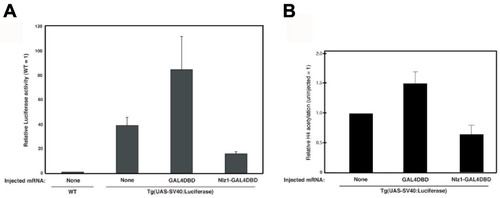
Nlz1 represses expression of the luciferase transgene and promotes histone deacetylation at the transgenic promoter in TG(UAS-SV40:Luciferase) embryos.A, B. TG(UAS-SV40:Luciferase) embryos were injected with mRNA encoding GAL4DBD or Nlz1-GAL4DBD as indicated under the graph. Embryos were raised for 12 hours and assayed for luciferase activity (A) or for histone H4 acetylation at the transgenic promoter (B) as described in the Methods section. Data are expressed as fold luciferase activity (A), or fold H4 acetylation (B) relative to un-injected control. A T-test indicates p < 0.0005 for the difference in acetylation level between GAL4DBD- and Nlz1GAL4DBD-injected embryos. EXPRESSION / LABELING:
|
|
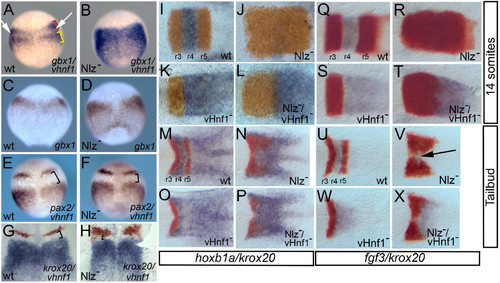
Nlz proteins act as repressors during zebrafish hindbrain development.A-H. Nlz proteins repress vhnf1 and gbx1 expression in rhombomere 4. Wild type embryos (A, C, E, G) or embryos injected with antisense morpholino oligos targeting Nlz1 and Nlz2 (Nlz-; B, D, F, H) were assayed by whole mount in situ hybridization for expression of gbx1/vhnf1 (A, B: gbx1 red bracket, vhnf1 yellow bracket), gbx1 (C, D), pax2/vhnf1 (E, F; both detected in purple) and krox20/vhnf1 (G, H; krox20 detected in red, vhnf1detected in blue). White arrows in A and black brackets in E-H indicate gaps in gene expression. I-X. Removing vhnf1 function restores late gene expression to Nlz- embryos. Wild type (I, M, Q, U), embryos injected with anti-Nlz MO (Nlz-; J, N, R, V), vhnf1 mutant embryos (K, O, S, W) and vhnf1 mutant embryos injected with anti-Nlz MO (Nlz-/vhnf1-; L, P, T, X) were assayed by in situ hybridization for expression of krox20 /hoxb1a (I-P; krox20 detected in red, hoxb1a detected in blue) or krox20/fgf3 (Q-X; krox20 detected in red, fgf3 detected in blue) at the 14 somite stage (I-L; Q-T) or the tailbud stage (M-P; U-X). Embryos in I-X were dissected and flat-mounted such that only the hindbrain is shown. Anterior is to the top in A-H and to the left in I-X). EXPRESSION / LABELING:
PHENOTYPE:
|