- Title
-
Defective Excitatory/Inhibitory Synaptic Balance and Increased Neuron Apoptosis in a Zebrafish Model of Dravet Syndrome
- Authors
- Brenet, A., Hassan-Abdi, R., Somkhit, J., Yanicostas, C., Soussi-Yanicostas, N.
- Source
- Full text @ Cells
|
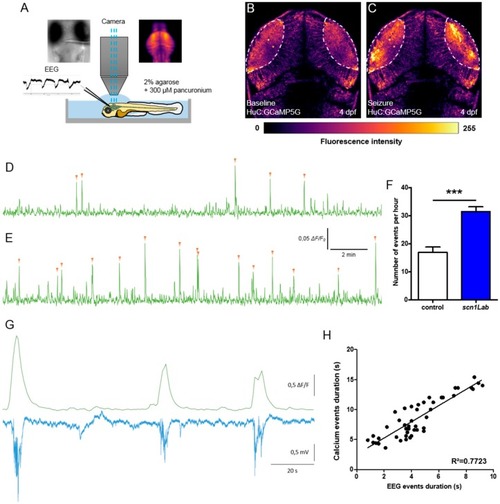
Correlation between LFP and calcium activity in PHENOTYPE:
|
|
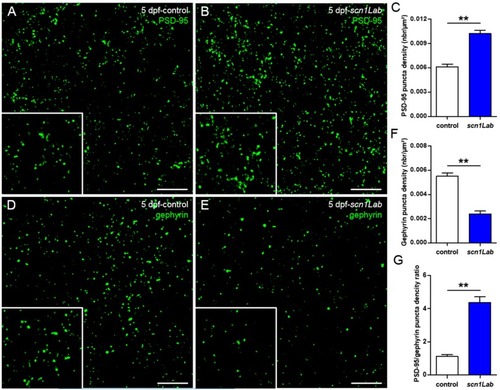
Defects of excitatory/inhibitory balance in the Scn1Lab-depleted larvae. ( |
|
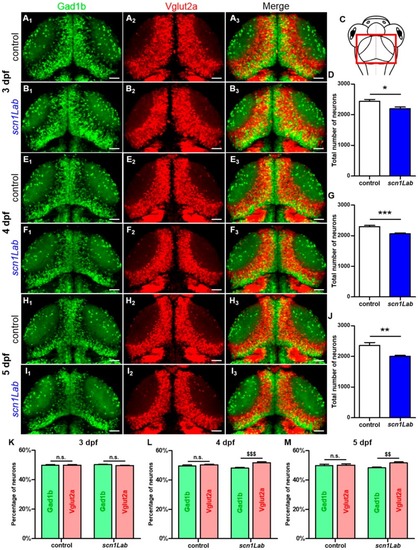
Evolution of the excitatory-inhibitory neuronal population in the EXPRESSION / LABELING:
PHENOTYPE:
|
|
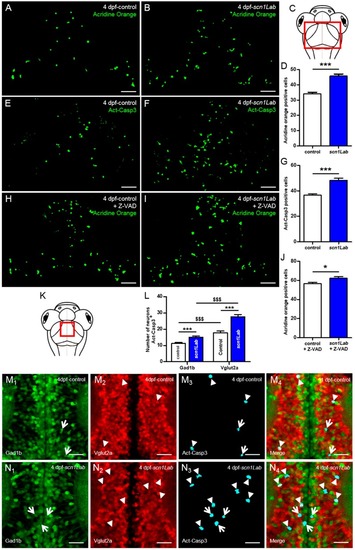
Increased neuronal death in Scn1Lab-depleted larvae. ( PHENOTYPE:
|