- Title
-
Regulation of neurocoel morphogenesis by Pard6gammab
- Authors
- Munson, C., Huisken, J., Bit-Avragim, N., Kuo, T., Dong, P.D., Ober, E.A., Verkade, H., Abdelilah-Seyfried, S., and Stainier, D.Y.
- Source
- Full text @ Dev. Biol.
|
s441 mutants show defects in the development of organs derived from epithelial tissues. (A, B) Comparison of 36 hpf wildtype and s441 mutant embryos. (B) s441 mutant embryos show defects in heart development (red arrow), failure of the brain ventricles to inflate (black arrow), and dorsal curvature. (C, D) Dorsal views comparing 30 hpf wildtype and s441 mutant hearts as assessed by myl7 expression. (C) In wildtype embryos, the heart elongates to form a tube. (D) In s441 mutant embryos, the heart fails to elongate. (E, F) Transverse sections of 72 hpf wildtype and s441 mutant larvae through a region just posterior to the endodermal organ-forming region. The pronephric ducts marked by acetylated-tubulin staining are also visible in these sections (white arrowheads). (E) Wildtype larvae form one polarized lumen in the posterior gut (red arrow). (F) s441 mutants occasionally show multiple lumens in the gut (red arrows). (G, H) Transverse sections of 30 hpf Tg(gutGFP)s854 wildtype and s441 mutant embryos. Sections are through the endodermal organ-forming region and were stained for filamentous actin and aPKC. (G) In wildtype embryos, the right side of the LPM (outlined in white) moves ventrally towards the endoderm while the left side (outlined in yellow) moves dorsally. (H) In s441 mutant embryos, the LPM fails to undergo asymmetric morphogenesis and both sides move dorsal to the endoderm. (I, J) Rhodamine dextran injected ventricles. Lateral (I) and dorsal (I′) views show that wildtype embryos have well-inflated (arrow) and continuous (arrowhead) brain ventricles. Lateral (J) and dorsal (J′) views illustrate that s441 mutant embryos do not form fully inflated brain ventricles (arrow) and that the lumen is discontinuous (arrowhead). (K, K′) Lateral views of 32 hpf wildtype and s441 mutant embryos show the absence of an inflated hindbrain ventricle (arrows) in mutants compared to wildtype. EXPRESSION / LABELING:
PHENOTYPE:
|
|
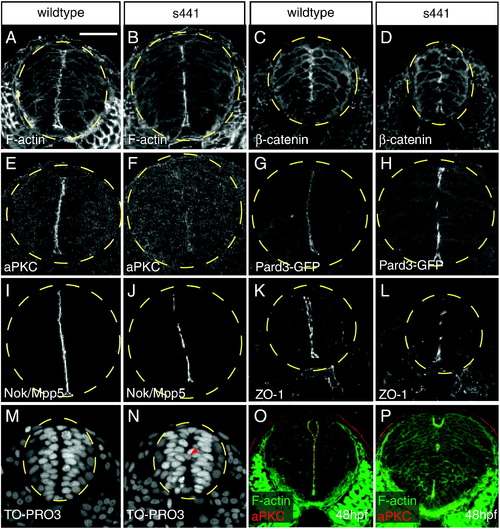
s441 mutants fail to form continuous apical membranes in the neural tube. (A-P) Transverse sections through wildtype and s441 mutant neural tubes between the first and eighth somite at 24 (A?N) and 48 hpf (O?P). The dashed yellow circles outline the neural tube. Embryos were analyzed for localization of filamentous actin (A, B), β-catenin (C, D), aPKC (E, F), Pard3:GFP (G, H), Nok/Mpp5 (I?J), ZO-1 (K, L), TO-PRO3 (M, N), and both filamentous actin and aPKC (O?P). (A, C, E, G, I, K) Wildtype embryos form continuous apical membranes. (B, D, F, H, J, L) s441 mutant embryos form discontinuous apical membranes. (F) Note that apical aPKC levels are dramatically reduced in s441 mutants. (M) In wildtype neural tubes, nuclei are organized and positioned away from the midline. (N) In s441 mutant neural tubes, some nuclei (arrowhead) are positioned across the midline. (panels O?P) At 48 hpf, the apical membranes in the s441 mutant neural tubes remain discontinuous. Scale bar represents 50 μm and is applicable to all panels in this figure. |
|
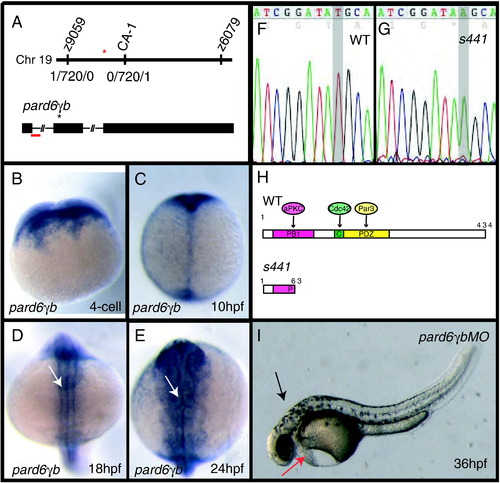
s441 is a mutation in pard6γb. (A) Positional cloning of s441. s441 was linked to a region on Chromosome 19 between the SSLP markers z9059 and z6079. Using an additional SSLP marker (labeled CA-1) the position of the mutation was narrowed to a region flanked by two recombinants, each unique to one side. The red asterisk represents the location of pard6γb. pard6γb contains three exons (rectangles) and two introns (lines). The black asterisk represents the lesion. The red bar represents the region targeted by the splice-blocking morpholino. (B?E) Expression of pard6γb. (B) pard6γb mRNA is maternally deposited. (C) At 10 hpf, pard6γb appears to be expressed ubiquitously. (D, E) At 18 and 24 hpf, pard6γb appears to be expressed in all tissues and shows heightened expression in the neural tube (arrows). (F, G) Sequence analysis of wildtype versus s441 pard6γb cDNA revealed a T to A (gray highlight in panels H and I) transition 192 base pairs following the start of translation. (H) Schematic of the PB1, CRIB (abbreviated C), and PDZ domains and the predicted protein-protein interactions with the wildtype protein. The premature stop codon caused by the s441 mutation leads to a protein truncated at amino acid 63. Numbers shown represent amino acid position. (I) Injecting 2 ng of a splice morpholino designed against the first exon-intron boundary phenocopied the s441 mutation. The morphants showed defects in heart development (red arrow), failure of the brain ventricles to inflate (black arrow), and dorsal curvature. |
|
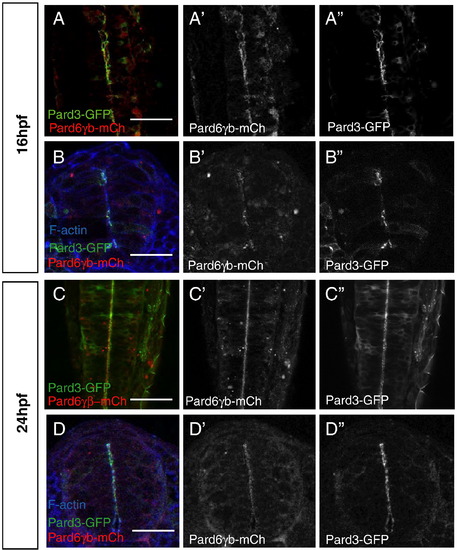
Distribution of Pard6γb during neurulation. (A?D) All embryos were injected with pard3-gfp and pard6γb-mch mRNA. (A, C) Optical sections through the neural tube of live embryos (dorsal views between the first and sixth somite, anterior up). (B, D) Transverse sections through the neural tube of embryos between the first and sixth somite stained for filamentous actin. (A, B) At 16 hpf, Pard6γb-mCh begins to localize with Pard3-GFP to the forming apical membranes of the neural tube. (C, D) At 24 hpf, Pard6γb-mCh localizes with Pard3-GFP along the maturing apical membranes of the neural tube. Scale bars represent 50 μm. |
|
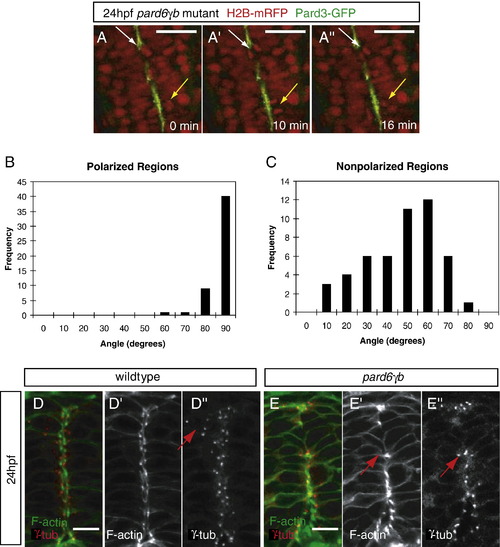
Position of the apical membranes in the neural tube appears to regulate the orientation of mitoses and the positioning of centrosomes during the neural tube stage. (A) Time lapse images of a 24 hpf pard6γbs441 mutant embryo injected with pard3-gfp and h2b-mrfp mRNA (dorsal views between the first and sixth somite, anterior to the top). Time is indicated in minutes. The yellow arrow points to a cell division in a region of continuous polarity, at an 88° angle (see B). The white arrow points to a cell division in a region of discontinuous polarity, at a 26° angle (see panel C). (B, C) Schematic of the technique used to analyze the angle of cell division in wildtype and pard6γbs441 mutant embryos. The angle between the midline (green) of the neural tube and the cleavage plane (black) of two dividing nuclei was recorded and analyzed on a histogram using the value ≤ 90°. (B) In regions with polarized localization of Pard3-GFP in wildtype (n = 3) and pard6γbs441 mutant (n = 12) embryos, the angle of division clustered between 80° and 90° (n = 40/50). (C) In regions lacking polarized localization of Pard3-GFP in pard6γbs441 mutant embryos, the angle of division clustered between 40° and 70° (n = 29/50). (D, E) Transverse sections through the neural tube between the first and sixth somite of 24 hpf embryos, stained for filamentous actin and γ-Tubulin. (D) In wildtype embryos, centrosomes are localized along the apical side of the cells lining the neurocoel or rotated 90° when cells are dividing (arrow). (panel F) In pard6γbs441 mutant embryos, centrosomes are positioned as in wildtype when polarized apical membranes are present, but appear to be missing or mislocalized in nonpolarized regions. The arrow points to a cell that has re-oriented the position of its centrosome towards a polarized region. Scale bars in panel A represent 50 μm, and in panels D and E 10 μm. |
|
The PB1, CRIB and PDZ domains are required for Pard6γb function but not localization. (A?E) Live, 24 hpf embryos injected with h2b-mRFP and pard6γb(Δpb1)-GFP (abbreviated ΔPB1-GFP), pard6γb(Δcrib)-GFP (abbreviated ΔCRIB-GFP) or pard6γb(kplg)-GFP (abbreviated KPLG-GFP) mRNA as indicated (dorsal views between the first and sixth somite, anterior to the top). (A?C) In wildtype neural tubes, nuclei are organized on either side of the midline. (D-F) In pard6γbs441 mutant neural tubes, nuclei are disorganized. In wildtype embryos, ΔPB1-GFP (A), ΔCRIB-GFP (B) and KPLG-GFP (C) localized to the cytoplasm and apical membranes. In pard6γbs441 mutant embryos, ΔPB1-GFP (D), ΔCRIB-GFP (E) and KPLG-GFP (F) failed to rescue the mutant phenotype and localized to the discontinuous apical membranes. Scale bars represent 50 μm. |
|
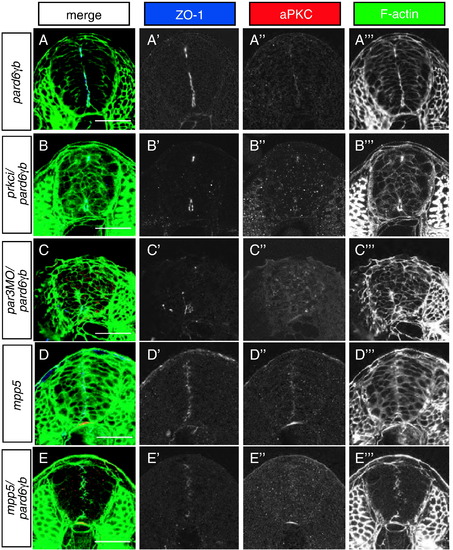
Role of the Par3/Par6/aPKC complex in apical membrane formation in the neural tube. All images represent transverse sections between the first and sixth somite stained for ZO-1 (blue), aPKC (red), and filamentous actin (green) at 24 hpf. (A) pard6γbs441 mutant neural tubes show a reduced amount of apical membranes. (B) pard6γbs441/prkci double mutants show a more severe reduction in apical membrane formation than pard6γbs441 mutants. (C) pard6γbs441 mutants injected with a pard3 morpholino show a more severe reduction in apical membrane formation than pard6γbs441 mutants. (D) mpp5 mutant neural tubes do not show an obvious severe reduction in apical membrane formation. (E) pard6γbs441/mpp5 double mutants show a more severe reduction in apical membrane formation than pard6γbs441 or mpp5 single mutants. In addition, pard6γbs441/prkci and pard6γbs441/mpp5 double mutants as well as pard6γbs441 mutants injected with a pard3 morpholino exhibit a severe epithelial disorganization in the neural tube. Scale bars represent 50 μm. |
|
Apical localization of the other Pard6 family members and partial rescue of the pard6γbs441 mutant phenotype by Pard6α. (A, C, E, G, I, K) Optical sections through the dorsal neural tube of live 24 hpf embryos between the first and sixth somite. (B, D, F, H, J, L) Transverse sections through the neural tube between the first and sixth somite of 24 hpf embryos, stained for filamentous actin. (A?D) Embryos injected with mRNA encoding Pard6γa-GFP. (A, B) In wildtype embryos, Pard6γa-GFP localized to the apical membranes. (panels C?D) In pard6γbs441 mutant embryos, Pard6γa-GFP localized to the discontinuous apical membranes, and it did not rescue the s441 mutant phenotype. (E?H) Embryos injected with mRNA encoding Pard6β-mCh. (E, F) In wildtype embryos, Pard6β-mCh localized to the apical membranes. (G, H) in pard6γbs441 mutant embryos, Pard6β-mCh localized to the discontinuous apical membranes, and it did not rescue the pard6γbs441 mutant phenotype. (I?L) Embryos injected with mRNA encoding Pard6α-GFP. (I, J) In wildtype embryos, Pard6α-GFP localized to the apical membranes. (K, L) In pard6γbs441 mutant embryos, Pard6α-GFP localized to the apical membranes. (K) In this pard6γbs441 mutant embryo, midline Pard6α-GFP localization was discontinuous and Pard6α-GFP failed to rescue the neurocoel phenotype. (L) In another, and genotyped, pard6γbs441 mutant embryo, midline Pard6α-GFP localization was continuous and Pard6α-GFP rescued the neurocoel phenotype. Scale bars represent 50 μm. |
|
Localization of Pard6γb prior to 24 hpf. (A?D) Images of live wildtype embryos. Embryos were injected with pard3-gfp, pard6γb-mch, pard6γb-gfp, and/or h2b-mrfp mRNA as indicated. (A) At 10 hpf, Pard3-GFP localizes primarily to the cytoplasm and cell membranes. (B) At 10 hpf, Pard6γb-mCh localizes to the nucleus, cytoplasm and cell membranes. [No nuclear localization signal was found in the Pard6γb-GFP fusion protein]. (C) At 10 hpf, Pard6γb-GFP co-localizes with H2B-mRFP in the nucleus. (D) At 13 hpf, Pard6γb-GFP begins to change its localization pattern. It localizes primarily to the cytoplasm and cell membranes. |
|
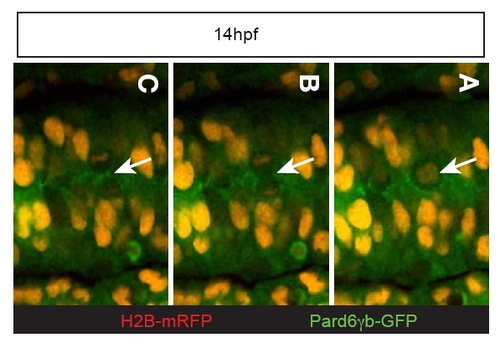
Distribution of Pard6γb during neurulation. (A?C) Mitotic event from an embryo injected with pard6γb-gfp and h2b-mrfp mRNA. (C) Pard6γb-GFP is initially all around the membrane and then appears to localize to the cleavage furrow during cytokinesis (arrow) at 14 hpf. |
|
Maternal Zygotic (MZ) pard6γbs441 mutant embryos. (A, A′) 36 hpf MZpard6γbs441 mutant embryo. (A) The overall body phenotype of maternal zygotic mutant embryos is similar to zygotic mutants. (A′) The retinal pigmented epithelium is more severely affected in maternal zygotic mutants. (B) A MZpard6γbs441 mutant embryo sectioned between the first and sixth somite and stained for ZO-1 (blue), aPKC (red), and filamentous actin (green) at 30 hpf. MZpard6γbs441 mutant embryos show a multiple lumen phenotype similar to that seen in zygotic pard6γbs441 mutants. PHENOTYPE:
|
|
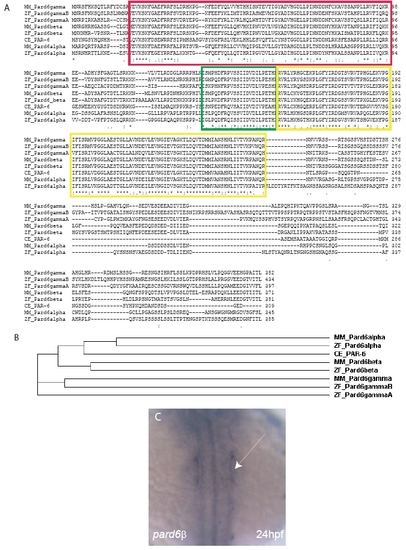
The Pard6 protein family. (A) Clustal W (1.83) Multiple Sequence Alignment of the four zebrafish (ZF) Pard6 proteins with the three mouse (MM) Pard6 proteins and the C.elegans (CE) PAR-6 protein. The red box outlines the conserved PB1 domain. The green box outlines the conserved CRIB domain. The yellow boxes outline the conserved PDZ domain. (B) Cladogram of the four zebrafish (ZF) Pard6 proteins, three mouse (MM) Pard6 proteins, and the C.elegans (CE) PAR-6 proteins. (C) in situ hybridization analysis shows pard6β expression at 24 hpf in the otic placodes (arrow). EXPRESSION / LABELING:
|
Reprinted from Developmental Biology, 324(1), Munson, C., Huisken, J., Bit-Avragim, N., Kuo, T., Dong, P.D., Ober, E.A., Verkade, H., Abdelilah-Seyfried, S., and Stainier, D.Y., Regulation of neurocoel morphogenesis by Pard6gammab, 41-54, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.