Fig. 6
- ID
- ZDB-FIG-250818-40
- Publication
- Kawasaki et al., 2025 - Meioc-Piwil1 complexes regulate rRNA transcription for differentiation of spermatogonial stem cells
- Other Figures
- All Figure Page
- Back to All Figure Page
|
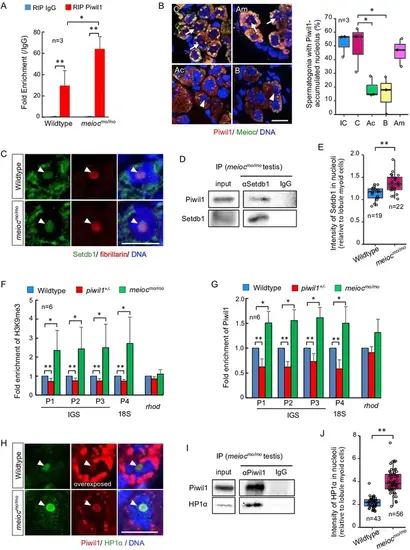
Nucleolar Piwil1 interacted with Setdb1 and caused silenced epigenetic state of rDNA loci. (A) Fold enrichment of pre-rRNA (5’ETS-18S rRNA) in Piwil1 immunoprecipitated RNA relative to the control IgG in wild-type and meiocmo/mo testes. (B) Immunostaining of Piwil1 (left panels) and the percentage of spermatogonia with detectable nucleolar Piwil1 (right panel) in the meiocmo/mo testes treated with α-amanitin (Am), actinomycin D (Ac), and BMH-21 (B). Arrows: Piwil1 detectable nucleoli; arrowheads: Piwil1 undetectable nucleoli; C: control without inhibitors; IC: initial control. (C) Immunostaining of Setdb1 and fibrillarin in wild-type and meiocmo/mo spermatogonia. Arrowheads: nucleoli. (D) Co-IP of Piwil1 and Setdb1 using meiocmo/mo testes lysate. Piwil1 was detected in Setdb1 IP. (E) Intensities of Setdb1 in nucleoli in wild-type and meiocmo/mo spermatogonia. (F, G) ChIP-qPCR analysis of H3K9me3 (F) and Piwil1 (G) levels in 45S-rDNA region in wild-type, piwil1+/-, and meiocmo/mo testes. The position of primers was indicated in Figure 2—figure supplement 2. Mean ± SD are indicated. (H) Immunostaining of HP1α and Piwil1 in wild-type and meiocmo/mo spermatogonia. Arrowheads: nucleolus. (I) Co-IP of Piwil1 and HP1α using meiocmo/mo testis lysate. HP1α was detected in Piwil1 IP. (J) Intensities of HP1α in nucleoli in wild-type and meiocmo/mo spermatogonia. *p<0.05, **p<0.01. Scale bars: 10 µm. |