Fig. 5
- ID
- ZDB-FIG-250807-55
- Publication
- Lush et al., 2025 - Stem and progenitor cell proliferation are independently regulated by cell type-specific cyclinD genes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
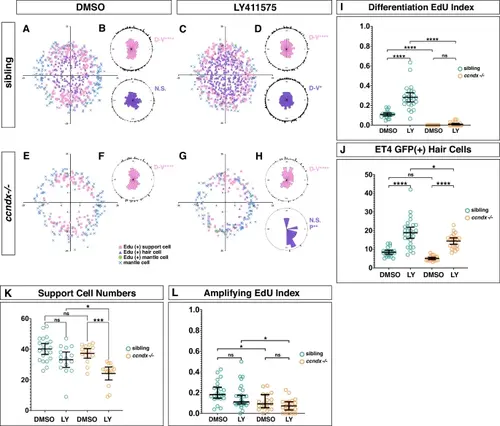
ccndx is required for the increased proliferation induced by Notch inhibition. A Spatial analysis of EdU+ support cells and hair cells after 24 h of regeneration in control DMSO treated sibling neuromasts. B One-sided binomial analysis of EdU+ cell positions. n = 21. ****p < 0.00001, ns=not significant, p = 0.17. C Spatial analysis of EdU+ support cells and hair cells after 24 h of regeneration in LY411575 treated sibling neuromasts. D One-sided binomial analysis of EdU+ cell positions. n = 27. ****p < 0.00001, **p < 0.02. E Spatial analysis of EdU+ support cells and hair cells after 24 hrs of regeneration in control DMSO treated ccndx−/− neuromasts. F One-sided binomial analysis of EdU+ cell positions. There are no EdU+ hair cells in ccndx−/− neuromasts. n = 18. ****p < 0.00001. G Spatial analysis of EdU+ support cells and hair cells after 24 h of regeneration in LY411475 treated ccndx−/− neuromasts. H One-sided binomial analysis of EdU+ cell positions. There are few EdU+ hair cells in LY411575 treated ccndx−/− neuromasts. n = 21. ****p < 0.00001, **p < 0.006, ns=not significant, p = 0.11. I EdU index of differentiation divisions after DMSO and LY411575 (LY) treatment in sibling and ccndx−/− regenerating neuromasts. There is no significant difference in differentiating EdU indexes between DMSO or LY411575 treated ccndx−/− neuromasts. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 1, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−. J Quantification of sqEt4:EGFP+ hair cells in DMSO and LY411575 (LY) treated sibling or ccndx−/− neuromasts after 24 h of regeneration. LY411575 induces an increase in regenerated hair cells in both siblings and ccndx−/− neuromasts. ns = not significant p = 0.18, ****p < 0.0001, *p = 0.0113, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−. K Quantification of support cell numbers in DMSO and LY411575 (LY) treated sibling or ccndx−/− neuromasts after 24 h of regeneration. LY411575 decreases support cells in both siblings and ccndx−/− neuromasts. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.067 and p = 0.84, *p < 0.024, ***p = 0.0007, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−. L EdU index of amplifying cells in DMSO and LY411575 (LY) treated sibling or ccndx−/− neuromasts after 24 h of regeneration. ns = not significant p = 0.46 and p = 0.78, *p < 0.018, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 21 for DMSO sibling, n = 27 for LY411575 sibling, n = 18 for DMSO ccndx−/− and n = 21 for LY411575 ccndx−/−. |