Fig. 3
- ID
- ZDB-FIG-250807-53
- Publication
- Lush et al., 2025 - Stem and progenitor cell proliferation are independently regulated by cell type-specific cyclinD genes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
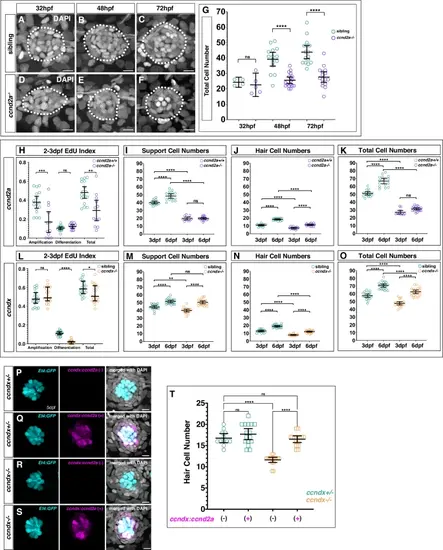
ccnd2a and ccndx are required for support cell and progenitor cell proliferation during development, respectively.A–F DAPI stained nuclei of neuromasts from sibling (A - C) or ccnd2a−/− (D - F) at 32, 48 or 72 hpf. Scale bar = 10 µm. G Quantification of total cell number from DAPI stained neuromasts in sibling and ccnd2a−/− at 32, 48 or 72 hpf. ns = not significant p = 0.98, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 4 for 32 hpf sibling, n = 5 for 32 hpf ccnd2a−/−, n = 15 for 48 hpf sibling, n = 18 for 48 hpf ccnd2a-/, n = 16 for 72 hpf sibling and i = 16 for 72 hpf ccnd2a−/−. H Comparison of wildtype and ccnd2a−/− amplifying, differentiation or total EdU indexes between 2 − 3 dpf. ccnd2a−/− has reduced amplification and total EdU indexes. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.55, ***p = 0.0008, **p = 0.0016, two-tailed t-test. n = 16 for ccnd2a+/+ and n = 12 for ccnd2a−/−. Comparisons of wildtype and ccnd2a−/− support- (I) hair cell- (J) and total (K) cell numbers. ns = not significant p = 1 for (I) and p = 0.087 (K), ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. Data are presented as mean values + /− 95% confidence intervals. n = 16 for 3dpf wildtype, n = 16 for 6dpf wildtype, n = 12 for 3dpf mutant and n = 20 for 6dpf mutant. L Comparison of sibling and ccndx−/− amplifying, differentiation or total EdU indexes between 2 − 3 dpf. Similarly to 5 − 6 dpf, ccndx−/− neuromasts show reduced differentiation and total EdU indexes and no change in the amplification index. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.25, ****p < 0.0001, *p = 0.0467, two-tailed t-test. n = 18 for both. M– O Comparisons of sibling and ccndx−/− support-, hair cell- and total cell numbers. Data are presented as mean values + /− 95% confidence intervals. ns=not significant p = 0.91, ****p < 0.0001, **p = 0.0019, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. P–S 5 dpf neuromasts from ccndx+/- (P, Q) or ccndx−/− (R, S) expressing sqEt4:GFP without or with ccnd2a-P2A-mScarlet-I driven by the ccndx promoter. Scale bar = 10 µm. T Quantification of sqEt4:GFP+ hair cells in ccndx+/- or ccndx−/− neuromasts at 5 dpf without or with ccnd2a-P2A-mScarlet-I. n = 12 for ccndx+/-/ccnd2a-P2A-mScarlet-I- and n = 16 for all other groups. Data are presented as mean values + /− 95% confidence intervals. ns = not significant p = 0.11 and p = 1, ****p < 0.0001, Two-way Anova (genotype, condition) with Šidák multiple comparisons test. |