Fig. 4
- ID
- ZDB-FIG-250416-45
- Publication
- Childers et al., 2025 - Protein absorption in the zebrafish gut is regulated by interactions between lysosome rich enterocytes and the microbiome
- Other Figures
- All Figure Page
- Back to All Figure Page
|
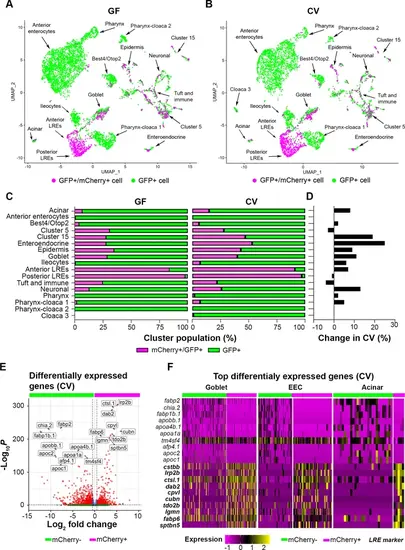
Uptake of mCherry occurs in cells enriched in lysosome-rich enterocyte (LRE) markers. (A, B) UMAP projections highlighting mCherry-positive/GFP-positive cells (magenta) and GFP-positive cells (green) in the germ-free (GF) and conventional (CV) datasets. (C) Bar plots portray the percentage of mCherry-positive and mCherry-negative cells in the GF and CV datasets. Bar color indicates the proportion of mCherry-positive (magenta) and mCherry-negative (green) cells in each cluster. (D) Bar plot displays the difference in the proportion of mCherry-positive cells in the CV compared to the GF dataset. Positive values show that the proportion of mCherry-positive cells were higher in the CV dataset. (E) Volcano plot shows differentially expressed genes between mCherry-positive and mCherry-negative cells in the CV dataset. The x-axis displays the log fold change in expression between mCherry-positive and mCherry-negative cells, with positive values showing enhanced expression in mCherry-positive cells and negative values showing higher expression in mCherry-negative cells. Red points are genes with significantly different expression (padj <0.05) and high fold change (log2FC < - 0.05, log2FC >0.05). (F) Heatmap displays the expression of the top markers for mCherry-positive and mCherry-negative cells in goblet, EEC, and acinar clusters. The color bar at the top indicates mCherry-positive (magenta) and mCherry-negative (green) cell types. Expression level is highlighted with a color gradient. mCherry-positive cells showed higher expression of dab2 and other LRE-enriched endocytic markers (bolded), whereas mCherry-negative cells express typical anterior enterocyte markers such as fabp2. |