|
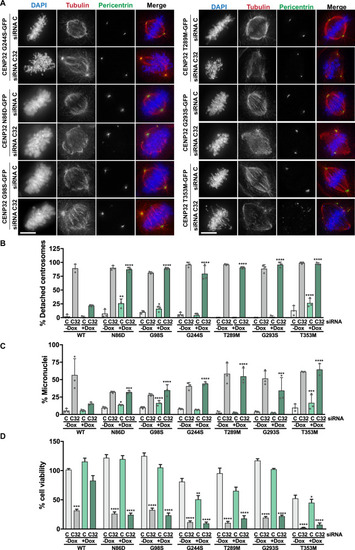
SPOUT1/CENP-32 patient variants lead to alterations of spindle organisation. A Representative fluorescence images of a Control (siRNA C) or SPOUT1/CENP-32 (siRNA C32) siRNA rescue assay in stable U2OS cell lines with inducible expression of different SPOUT1/CENP-32-GFP constructs. Conditions with doxycycline only; conditions without doxycycline in Fig. S17A. B, C Quantification for the analysis of the centrosome detachment phenotype (% of cells with detached centrosomes; B) and chromosome segregation errors (% of micronuclei; C) in inducible U2OS cell lines expressing SPOUT1/CENP-32 WT-GFP or the SPOUT1/CENP-32 patient variants. For easy direct comparison with the SPOUT1/CENP-32 variant conditions, the SPOUT1/CENP-32 WT conditions shown in Fig. 6D are also shown in this figure. Data are representative of a minimum of three biological replicates, n ≥ 70 cells analyzed in total per treatment, mean ± SD, n = 3. Scale bar, 10 μm. Two-sided Fisher’s exact test with Bonferroni correction (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). Exact p-values for the detachment centrosome phenotype (B) vs WT condition are: N86D siRNA C +Dox=3.1E-03, N86D siRNA C32 +Dox=4.3E-30, G98S siRNA C +Dox=1.9E-02, G98S siRNA C32 +Dox=4.9E-33, G244S siRNA C32 +Dox=4.8E-15, T289M siRNA C32 +Dox=1.8E-25, G293S siRNA C32 +Dox=2.7E-30, T353M siRNA C +Dox=3.3E-07, T353M siRNA C32 +Dox=4.1E-28. Exact p-values for the micronuclei phenotype (C) vs WT condition are: N86D siRNA C +Dox=1.3E-02, N86D siRNA C32 +Dox=2.9E-04, G98S siRNA C32 -Dox=1.9E-07, G98S siRNA C +Dox=2.5E-04, G98S siRNA C32 +Dox=2.1E-05, G244S siRNA C32 +Dox=1.6E-15, T289M siRNA C32 +Dox=1.2E-24, G293S siRNA C32 +Dox=1.3E-03, T353M siRNA C +Dox=1.1E-04, T353M siRNA C32 +Dox=5.5E-37. Data points for each of the replicates are shown. Source data are provided as a Source Data file. D Cell viability evaluation of inducible U2OS cell lines expressing SPOUT1/CENP-32 WT-GFP or SPOUT1/CENP-32 patient variants assessed by the MTT assay. For easy direct comparison with the SPOUT1/CENP-32 variant conditions, the SPOUT1/CENP-32 WT conditions shown in Fig. 6E are also shown in this figure. Three independent biological replicates, mean ± SEM. WT siRNA C -Dox: n = 26; WT siRNA C32 -Dox; n = 26; WT siRNA C +Dox: n = 25; WT siRNA C32 +Dox: n = 27; N86D siRNA C -Dox: n = 22; N86D siRNA C32 -Dox: n = 25; N86D siRNA C +Dox: n = 27; N86D siRNA C32 +Dox: n = 22; G98S siRNA C -Dox: n = 27; G98S siRNA C32 -Dox; n = 26; G98S siRNA C +Dox: n = 27; G98S siRNA C32 +Dox: n = 24; G244S siRNA C -Dox: n = 25; G244S siRNA C32 -Dox; n = 27; G244S siRNA C +Dox: n = 27; G244S siRNA C32 +Dox: n = 27; T289M siRNA C -Dox: n = 18; T289M siRNA C32 -Dox; n = 18; T289M siRNA C +Dox: n = 18; T289M siRNA C32 +Dox: n = 18; T353M siRNA C -Dox: n = 18; T353M siRNA C32 -Dox; n = 18; T353M siRNA C +Dox: n = 18; T353M siRNA C32 +Dox: n = 18; G293S siRNA C -Dox: n = 27; G293S siRNA C32 -Dox; n = 27; G293S siRNA C +Dox: n = 27; G293S siRNA C32 +Dox: n = 27. Non-parametric ANOVA with Kruskal-Wallis followed by Dunn’s multiple comparisons test (*, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001). Exact p-values vs WT siRNA C -Dox condition are: WT siRNA C32 -Dox=1.4E-04, N86D siRNA C32 -Dox=2.4E-06, N86D siRNA C32 +Dox=4.6E-06, G98S siRNA C32 -Dox=8.7E-05, G98S siRNA C32 +Dox=2E-07, G244S siRNA C32 -Dox=1E-07, G244S siRNA C +Dox= 6.8E-03, G244S siRNA C32 +Dox=1E-07, T289M siRNA C32 -Dox=1E-07, T289M siRNA C32 +Dox= 1E-07, G293S siRNA C32 -Dox=1E-07, G293S siRNA C32 +Dox=1E-07, T353M siRNA C32 -Dox= 1E-07, T353M siRNA C +Dox=1.4E-02, T353M siRNA C32 +Dox=1E-07. Source data are provided as a Source Data file.
|