|
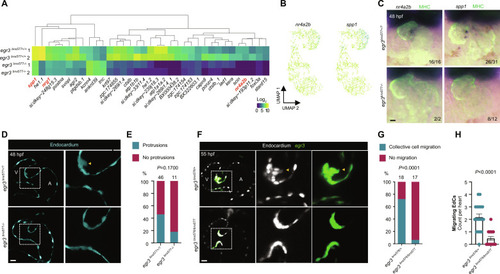
egr3 is necessary for EdC migration, which is required for cardiac valve formation. (A) RNA-seq heatmap analysis of differentially expressed genes; fold change ± 1.5, Padj < 0.05, mean > 5. (B) scRNA-seq endocardial expression pattern of Egr3 targets nr4a2b and spp1. (C) In situ hybridization for Egr3 targets nr4a2b and spp1 in egr3+/+ and egr3−/− siblings at 48 hpf. (D) Confocal images of representative hearts from 48 hpf egr3+/? and egr3−/− sibling embryos displaying, respectively, AV EdC protrusions toward the ECM and no protrusions; EdCs are marked by Tg(kdrl:eGFP) expression (cyan). (E) Percentage of hearts from 48 hpf egr3+/? and egr3−/− sibling embryos displaying AV EdC protrusions toward the ECM; three independent experiments. (F) Confocal images of representative hearts from 55 hpf egr3+/− and egr3−/− sibling embryos displaying, respectively, collective AV EdC migration and no migration; EdCs are marked by Tg(kdrl:NLS-mCherry) expression (gray) and AV EdCs by Pt(egr3:Gal4-VP16);Tg(UAS:eGFP) expression (green). (G) Percentage of hearts from 55 hpf egr3+/− and egr3−/− sibling embryos displaying AV EdC migration; three independent experiments. (H) Number of migrating AV EdCs per heart in 55 hpf egr3+/− and egr3−/− sibling embryos; n = 18 and 17 embryos; three independent experiments. Student’s t test, mean ± SEM. (E) and (G) Fisher’s exact test. Scale bars, 20 μm.
|